
Oral microbiome and risk of lung cancer: results from a two-sample mendelian randomization analysis
Highlight box
Key findings
• Oral microbiome, partly from saliva and tongue, is causally associated with lung cancer.
What is known and what is new?
• The oral microbiome may function as a biomarker for lung cancer screening.
• The oral microbiomes, Gemella haemolysans (pheno.388) from saliva, an unclassified species (pheno.844) of Clostridia from saliva and an unclassified species (pheno.1354) of Prevotell from tongue, were causally associated with lung cancer.
What is the implication, and what should change now?
• Oral microbiota is closely linked to lung cancer, indicating its potential as a noninvasive tool for early screening and diagnosis. Early screening of the oral microbiome in high-risk groups, combined with timely interventions, may reduce lung cancer risk. Regarding treatment, personalized therapy based on oral flora may provide novel therapeutic strategies for lung cancer.
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide, with two primary histopathologic types: non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC is the most common, accounting for approximately 80% to 85% of lung cancers. It includes adenocarcinoma (about 40% of NSCLC cases), squamous cell carcinoma (25% to 30%), large cell carcinoma (10% to 15%), and other less common subtypes (1). Lung cancer is characterized by a high incidence and mortality rate, insidious onset, challenges in early diagnosis, and low survival rates. Therefore, early diagnosis and treatment are crucial to improving overall survival (1). While existing markers for early screening, diagnosis, and prognostic assessment have limited sensitivity and specificity, the identification of more effective biomarkers could greatly benefit lung cancer patients.
The oral microbiome is highly diverse, comprising bacteria, fungi, viruses, archaea, and protozoa. Among them, the oral bacterial microbiome encompasses approximately 700 commonly occurring phylotypes (2). The 16S rDNA profiling of the healthy oral cavity categorized the inhabitant bacteria into six broad phyla, viz. Firmicutes, Actinobacteria, Proteobacteria, Fusobacteria, Bacteroidetes and Spirochaetes constitute 96% of total oral bacteria (3). Increasing evidence suggests a link between the oral microbiota and lung cancer development. Studies have demonstrated that lower alpha diversity in the oral microbiota is associated with a higher risk of lung cancer (4-6). On the one hand, the oral microbiome may undergo changes in lung cancer patients. For instance, Yan et al. observed significantly elevated levels of Capnocytophaga and Veillonella in patients with squamous cell carcinoma (SCC) of the lung (7). Similarly, Zhang et al. reported an increase in Veillonella and Streptococcus in the salivary microbiota samples of NSCLC patients (8). Cai and colleagues found family Fusobacteriaceae and Neisseriaceae, and order Bacteroidales in the mouth rinse samples of lung cancer patients (9). These three types of bacteria are among the most abundant in the lungs. On the other hand, alterations in the oral microbiome may influence lung cancer risk. Among the oral bacteria, order CW040 increases, while family Tissierellaceae and genus Parvimonas decrease the risk of lung cancer (9). Hosgood et al. showed that a greater abundance of the Bacilli class and Lactobacillales order in saliva correlates with a higher lung cancer risk, whereas increased levels of Spirochaetia and Bacteroidetes in saliva are associated with a lower risk (4). In summary, it can be hypothesized that lung cancer may be linked to a reduction in alpha diversity of oral bacteria, along with specific changes in bacterial abundance.
Therefore, we applied Mendelian randomization (MR) analysis to assess the causal relationship between the oral microbiome and lung cancer. Unlike randomized controlled trials (RCTs), MR excludes confounding factors and reverse causality by leveraging the principles of random allelic assignment and genetic variation from parents to offspring, respectively. We present this article in accordance with the STROBE-MR reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-2024-1163/rc).
Methods
Study design
This study utilized single nucleotide polymorphisms (SNPs) as genetic instruments to perform a two-sample MR analysis to evaluate the causal relationship between oral microbiome and lung cancer. The overall study framework is depicted in Figure 1. Assumption 1: association hypothesis, genetic instruments are reliably associated with exposure; Assumption 2: independence hypothesis, genetic instruments are not associated with confounding factors; Assumption 3: exclusivity hypothesis, genetic instruments are not independently associated with outcome, except through exposure. In this study, the oral microbiomes were considered to be the exposure, and lung cancer was the outcome. All analyses were based on publicly available summary data, and the original studies providing the data have received ethical approval. This study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments.
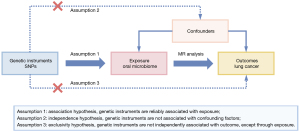
Exposure data
SNPs related to the human oral microbiome composition were selected as IVs from the large-scale metagenome-genome-wide association studies (mgGWAS) (10). The study included a cohort of 2,984 healthy individuals with 3,932 oral samples (2,017 tongue dorsum and 1,915 saliva) for high-depth whole metagenomic sequencing, to investigate the association between host genetics and oral microbiome composition.
Outcome data
The genome-wide association study (GWAS) summary statistics for lung cancer were obtained via the IEU-OpenGWAS online platform using query numbers of “ebi-a-GCST9001865” and “bbj-a-133”, including 4,444 cases and 174,282 controls of East Asian, and 4,050 cases and 208,403 controls of East Asian, respectively.
IVs
The following selection criteria were used to choose the optimal IVs: (I) potential IVs were selected based on SNPs significantly associated with the oral microbiome, with a significance threshold of P<5×10−8; (II) SNPs with an R2<0.001 and a clumping distance of 10,000 kb were excluded to account for linkage disequilibrium (LD); (III) SNPs having a minor allele frequency (MAF) of ≤0.01 were removed; (IV) palindromic SNPs (such as those with A/T or G/C alleles) were excluded to prevent errors due to strand orientation or allele coding issues; (V) the F-statistic is less than ten.
Statistical analysis
In this study, we performed a two-sample MR analysis to investigate whether there was a causal association between oral microbiome and lung cancer. The following two methods were employed to estimate causal effects: inverse variance weighted (IVW) and Wald ratio (11).
When a single SNP is used as an IV, the Wald ratio can be employed to estimate the causal effect by calculating the ratio of the SNP’s effect on the oral microbiome to its impact on lung cancer. If this ratio is significant and within a reasonable range, it may suggest a causal relationship between the oral microbiome and lung cancer.
When multiple SNPs are available (typically ≥2), the IVW method can be used. This method combines the estimates from individual SNPs using a meta-analysis approach to obtain a more robust estimate of the effect of the oral microbiome on cancer, thereby reducing the error introduced by relying on a single SNP. The IVW method assumes that all variants are valid IVs and that pleiotropy is balanced. When this assumption holds, the results are unbiased (12).
However, when there are too many IVs, ensuring the validity of each SNP is challenging. In such cases, additional methods like MR-Egger regression, weighted median, weighted mode, and simple mode can enhance the robustness of causal inference.
F-statistics were calculated to assess the strength of IVs in explaining the variance of the exposure variable (13). An F-value greater than 10 indicates that the instruments are sufficiently strong (13).
We considered the possibility of a bidirectional association, where lung cancer leads to changes in the oral microbiomes. To minimize the risk of this scenario, we performed MR Steiger’s test to strengthen the validity of the findings. When the P<0.5, it proves that there is no bidirectional association.
All statistical analyses were performed using the R version 4.2.1 and the R package TwoSampleMR (version 0.4.26).
Results
IVs
We obtained 1,549 oral microbiomes from saliva and 1,568 oral microbiomes from the tongue in the exposure data. Under the selection criteria for SNPs, we screened 52 SNPs from saliva and 68 SNPs from tongue. The related excluding SNPs were described in the method part and Tables S1,S2.
Association between oral microbiomes and lung cancer
A total of three oral microbiomes were causally associated with lung cancer in the Wald ratio or IVW model (Figures 2-5), namely Gemella haemolysans (pheno.388), an unclassified species (pheno.844) of Clostridia and an unclassified species (pheno.1354) of Prevotella. Unclassified species will be replaced with ID numbers in the analysis of results below. Gemella haemolysans [odds ratio (OR): 0.61, 95% confidence interval (CI): 0.42–0.88, P=0.009] in saliva was causally associated with lung cancer when the query number was “bbj-a-133”, while Gemella haemolysans (OR: 0.65, 95% CI: 0.45–0.93, P=0.02) was also causally associated with lung cancer when the query number was “ebi-a-GCST90018655”. Pheno.844 (OR: 0.61, 95% CI: 0.43–0.88, P=0.008) from saliva was causally associated with lung cancer in query number “bbj-a-133”. Pheno.1354 (OR: 1.47, 95% CI: 1.02–2.12, P=0.04) from tongue was causally associated with lung cancer in query number “bbj-a-133”. Among them, Gemella haemolysans and pheno.844 were protective factors for lung cancer (OR <1) and pheno.1354 was a risk factor for lung cancer (OR >1).
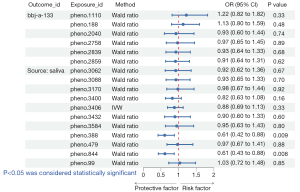
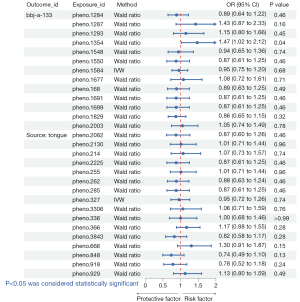
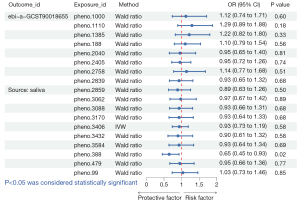
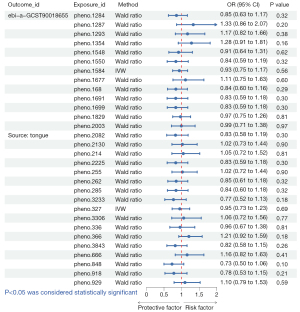
In the MR Steiger’s test, the P value of Gemella haemolysans was 2.83×10−8 when the query number was “ebi-a-GCST90018655” and 2.81×10−8 when the query number was “bbj-a-133”. Pheno.1354 had a P value of 1.40×10−8 and pheno.844 had a P value of 4.56×10−9. It demonstrates that there is no bidirectional association of causality between oral microbiomes and lung cancer.
Discussion
In this study, we performed two-sample MR analyses to investigate the causal association between oral microbiomes and lung cancer. The results showed that three oral microbiomes were causally associated with lung cancer. Gemella haemolysans and pheno.844 were protective factors for lung cancer, and pheno.1354 was a risk factor for lung cancer.
A growing body of research has demonstrated a correlation between the oral microbiome and lung cancer (4,5,8,9,14,15). Periodontal disease patients are linked to an increased risk of lung cancer (16,17). Numerous studies have shown that the saliva of healthy subjects contained a higher proportion of Gemella haemolysans than that of patients with periodontitis (18-21). It is hypothesised that Gemella haemolysans may be a protective factor against periodontal disease and lung cancer. This is consistent with our results, which showed a causal relationship between Gemella haemolysans and reduced lung cancer risk in both GWAS summary statistics with query numbers “ebi-a-GCST9001865” and “bbj-a-133”. Growth inhibition assays indicated that the protein components contained in the culture supernatant of Gemella haemolysans directly suppressed the growth of Porphyromonas gingivalis, correlating this effect with oral health (21). Periodontal disease is associated with specific microorganisms, including Porphyromonas gingivalis, and may also contribute to lung cancer risk through mechanisms such as dysbiosis (17,22,23). However, there is a lack of sufficient studies to elucidate in detail the mechanism of the relationship between Gemella haemolysans and periodontal disease and lung cancer. Another microbiome (id pheno.844) in our study that is a protective factor against lung cancer is a species of Clostridia, whose genus and species have not yet been classified. But studies suggested that the probiotic Clostridium butyricum, which is also a type of Clostridia, may have a positive impact on lung cancer patients receiving immunotherapy (24,25). What’s more, our results showed that the risk factor for lung cancer was an unclassified species (pheno.1354) of Prevotella. However, the oral microbiomes Prevotella intermedia, Prevotella nigrescens and Prevotella gingivalis were found to be positively associated with lung cancer (26). It is worth mentioning that the association mechanism between oral microbiomes and lung cancer is relatively complex and has not been fully identified so far.
Oral microbiota has been associated with a variety of diseases, including lung cancer. While smoking is the primary risk factor for lung cancer, the development of the disease is a complex process, and its causative factors, particularly the etiology of lung cancer in nonsmokers, remain inadequately clarified. A study involving nonsmoking female patients with NSCLC analyzed the salivary microbiota of 247 subjects, revealing that the diversity and abundance of oral microorganisms were significantly lower in cancer patients compared to healthy controls (5). In addition, a review summarized the association between oral microbiota and various cancers, stating that oral microorganisms such as Porphyromonas gingivalis, F. nuclearum, Streptococcus anginosus, Treponema denticola, and others mediate pro-inflammatory cytokine secretion, immunosuppression, apoptosis, and other mechanisms that promote cancer development (27). This suggests that the ecology of oral flora may be a new direction to explore the etiology of lung cancer in nonsmokers. A study found significant changes in the composition of the gut microbiota in lung cancer patients compared to the healthy population through 16S ribosomal RNA (rRNA) gene sequencing analysis and also found that a predictive model with 13 operational taxonomic unit (OTU)-based biomarkers achieved high accuracy in lung cancer prediction (28). As previously mentioned, oral microbiota is closely linked to lung cancer, indicating its potential as a noninvasive tool for early screening and diagnosis. Early screening of the oral microbiome in high-risk groups, combined with timely interventions, may reduce lung cancer risk. Regarding treatment, personalized therapy based on oral flora may provide novel therapeutic strategies for lung cancer. Some studies suggested that Clostridium butyricum may positively affect lung cancer patients receiving immunotherapy (24,25). Therefore, in terms of lung cancer prognosis, the oral microbiome may also serve as a biomarker of response for targeted therapy, immunotherapy, and postoperative recurrence, thereby facilitating the assessment and determination of patient response to these treatments.
It is equally important to acknowledge the limitations of our study. Firstly, the microbiome is an exposure phenotype with limited explanatory power, which means that robust calculations of the statistical efficacy of MR may be overly stringent. Second, since MR analyses are based on untestable assumptions, further experimental and clinical validation studies are essential to test the clinical significance of specific microbial species. In addition, there are still some limitations to this study, as stated below: (I) all three oral microbiomes causally associated with lung cancer had only one SNP, resulting in this study not being able to demonstrate the robustness of the results through sensitivity analyses. (II) Given the multiple comparisons across various oral microbiota and lung cancer subtypes, there is a risk of Type I errors. We did not apply strict multiple testing corrections (e.g., Bonferroni).
Conclusions
The oral microbiomes, Gemella haemolysans (pheno.388) from saliva, an unclassified species (pheno.844) of Clostridia from saliva and an unclassified species (pheno.1354) of Prevotella from tongue, were causally associated with lung cancer. Oral microbiology holds significant potential for clinical applications in etiologic exploration, early screening, prevention, and enhancing survival in lung cancer. Regarding treatment, personalized therapy based on oral flora may provide novel therapeutic strategies for lung cancer.
Acknowledgments
The authors wish to acknowledge the authors and participants of the involved GWAS for providing summary statistics data.
Footnote
Reporting Checklist: The authors have completed the STROBE-MR reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-2024-1163/rc
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-2024-1163/prf
Funding: None.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-2024-1163/coif). S.L. serves as an unpaid editorial board member of Translational Lung Cancer Research from February 2025 to January 2026. The other authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Schabath MB, Cote ML. Cancer Progress and Priorities: Lung Cancer. Cancer Epidemiol Biomarkers Prev 2019;28:1563-79. [Crossref] [PubMed]
- Palmer RJ Jr. Composition and development of oral bacterial communities. Periodontol 2000 2014;64:20-39. [Crossref] [PubMed]
- Verma D, Garg PK, Dubey AK. Insights into the human oral microbiome. Arch Microbiol 2018;200:525-40. [Crossref] [PubMed]
- Hosgood HD, Cai Q, Hua X, et al. Variation in oral microbiome is associated with future risk of lung cancer among never-smokers. Thorax 2021;76:256-63. [Crossref] [PubMed]
- Yang J, Mu X, Wang Y, et al. Dysbiosis of the Salivary Microbiome Is Associated With Non-smoking Female Lung Cancer and Correlated With Immunocytochemistry Markers. Front Oncol 2018;8:520. [Crossref] [PubMed]
- Zhang K, He C, Qiu Y, et al. Association of oral microbiota and periodontal disease with lung cancer: a systematic review and meta-analysis. J Evid Based Dent Pract 2023;23:101897. [Crossref] [PubMed]
- Yan X, Yang M, Liu J, et al. Discovery and validation of potential bacterial biomarkers for lung cancer. Am J Cancer Res 2015;5:3111-22.
- Zhang W, Luo J, Dong X, et al. Salivary Microbial Dysbiosis is Associated with Systemic Inflammatory Markers and Predicted Oral Metabolites in Non-Small Cell Lung Cancer Patients. J Cancer 2019;10:1651-62. [Crossref] [PubMed]
- Shi J, Yang Y, Xie H, et al. Association of oral microbiota with lung cancer risk in a low-income population in the Southeastern USA. Cancer Causes Control 2021;32:1423-32. [Crossref] [PubMed]
- Liu X, Tong X, Zhu J, et al. Metagenome-genome-wide association studies reveal human genetic impact on the oral microbiome. Cell Discov 2021;7:117. [Crossref] [PubMed]
- Gudicha DW, Schmittmann VD, Vermunt JK. Statistical power of likelihood ratio and Wald tests in latent class models with covariates. Behav Res Methods 2017;49:1824-37. [Crossref] [PubMed]
- Burgess S, Davey Smith G, Davies NM, et al. Guidelines for performing Mendelian randomization investigations: update for summer 2023. Wellcome Open Res 2019;4:186. [Crossref] [PubMed]
- Burgess S, Thompson SG. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol 2011;40:755-64. [Crossref] [PubMed]
- Hosgood HD 3rd, Sapkota AR, Rothman N, et al. The potential role of lung microbiota in lung cancer attributed to household coal burning exposures. Environ Mol Mutagen 2014;55:643-51. [Crossref] [PubMed]
- Cameron SJS, Lewis KE, Huws SA, et al. A pilot study using metagenomic sequencing of the sputum microbiome suggests potential bacterial biomarkers for lung cancer. PLoS One 2017;12:e0177062. [Crossref] [PubMed]
- Zeng XT, Xia LY, Zhang YG, et al. Periodontal Disease and Incident Lung Cancer Risk: A Meta-Analysis of Cohort Studies. J Periodontol 2016;87:1158-64. [Crossref] [PubMed]
- Yoon HS, Wen W, Long J, et al. Association of oral health with lung cancer risk in a low-income population of African Americans and European Americans in the Southeastern United States. Lung Cancer 2019;127:90-5. [Crossref] [PubMed]
- Arredondo A, Blanc V, Mor C, et al. Tetracycline and multidrug resistance in the oral microbiota: differences between healthy subjects and patients with periodontitis in Spain. J Oral Microbiol 2020;13:1847431. [Crossref] [PubMed]
- Kirst ME, Li EC, Alfant B, et al. Dysbiosis and alterations in predicted functions of the subgingival microbiome in chronic periodontitis. Appl Environ Microbiol 2015;81:783-93. [Crossref] [PubMed]
- Colombo AP, Bennet S, Cotton SL, et al. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. J Periodontol 2012;83:1279-87. [Crossref] [PubMed]
- Miyoshi T, Oge S, Nakata S, et al. Gemella haemolysans inhibits the growth of the periodontal pathogen Porphyromonas gingivalis. Sci Rep 2021;11:11742. [Crossref] [PubMed]
- Michaud DS, Liu Y, Meyer M, et al. Periodontal disease, tooth loss, and cancer risk in male health professionals: a prospective cohort study. Lancet Oncol 2008;9:550-8. [Crossref] [PubMed]
- Dong J, Li W, Wang Q, et al. Relationships Between Oral Microecosystem and Respiratory Diseases. Front Mol Biosci 2021;8:718222. [Crossref] [PubMed]
- Tomita Y, Ikeda T, Sakata S, et al. Association of Probiotic Clostridium butyricum Therapy with Survival and Response to Immune Checkpoint Blockade in Patients with Lung Cancer. Cancer Immunol Res 2020;8:1236-42. [Crossref] [PubMed]
- Tomita Y, Goto Y, Sakata S, et al. Clostridium butyricum therapy restores the decreased efficacy of immune checkpoint blockade in lung cancer patients receiving proton pump inhibitors. Oncoimmunology 2022;11:2081010. [Crossref] [PubMed]
- Zhou B, Lu J, Beck JD, et al. Periodontal and Other Oral Bacteria and Risk of Lung Cancer in the Atherosclerosis Risk in Communities (ARIC) Study. Cancer Epidemiol Biomarkers Prev 2023;32:505-15. [Crossref] [PubMed]
- Stasiewicz M, Karpiński TM. The oral microbiota and its role in carcinogenesis. Semin Cancer Biol 2022;86:633-42. [Crossref] [PubMed]
- Zheng Y, Fang Z, Xue Y, et al. Specific gut microbiome signature predicts the early-stage lung cancer. Gut Microbes 2020;11:1030-42. [Crossref] [PubMed]


