
Different prognosis of multiple lung cancer identified by 116-gene panel by next-generation sequencing based on an Asian population
Highlight box
Key findings
• Next-generation sequencing (NGS)-based 116-gene panel classification can improve the accuracy of diagnosing multiple primary lung cancer (MPLC) and intrapulmonary metastases (IPM). The diagnosis of IPM was associated with poor prognosis in Asian population.
What is known and what is new?
• Patients with multiple lung cancer are becoming more common. However, the optimal criterion to distinguish MPLC from IPM is still unclear. And the different diagnosis of multiple lung cancer may be associated with different prognosis in Asian population.
• To the best of our knowledge, this is the first study to report the different prognosis of multiple lung cancer identified by large-panel NGS in Asian population.
What is the implication, and what should change now?
• NGS-based 116-gene panel classification can help to diagnose IPM, which was associated with poor prognosis in Asian population. Large panel NGS may be considered as the routine examination for multiple lung cancer. For the treatment of the patients with diagnosis of IPM, we should consider adding more adjuvant therapies.
Introduction
Lung adenocarcinoma (ADC) has become the most common pathological type of lung cancer worldwide in recent years (1). The incidence of multiple nodules is increasing clinically, and multiple primary lung cancer (MPLC) accounts for 1–7% of cases (2,3). Lung ADC is characterized by significant histological and molecular heterogeneity. Distinguishing between MPLC and intrapulmonary metastases (IPM) is very difficult but has important implications for lung cancer diagnosis and treatment. MPLC nodules are staged individually, whereas IPM is staged as pathological T3 if nodules are in the same lobe, pathological T4 if nodules are in an ipsilateral lobe, or pathological M1a if nodules are in a contralateral lobe. When using the Martini and Melamed (MM) criteria, which is widely used clinically, it is difficult to diagnose MPLC correctly (4). Recent studies using molecular tools have highlighted significant limitations of the MM criteria. It is necessary to comprehensively consider all available clinical, radiological, pathological, and molecular information to distinguish MPLC from IPM, but the optimal criteria are unclear (5-7).
Over the years, more studies have begun to use molecular methods to identify MPLC and IPM, providing valuable information on the clonal relationship of lung ADC (8,9). In recent years, major driver gene mutations in two to five and 50-gene panel next-generation sequencing (NGS) have been performed to discriminate between MPLC and IPM. Although they provide significant information on the clonal relationship of lung ADCs, they also have some limitations (10-15).
In this study, we performed 116-gene panel NGS to molecularly classify MPLC and IPM. Our aim was to evaluate the value of broad panel NGS in the discrimination of lung cancer clonal relationships in clinical practice. We present this article in accordance with the STROBE reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-2024-1160/rc).
Methods
Patients
A total of 2,523 consecutive patients with lung cancer who underwent surgical resection in three thoracic departments of three medical centers (Fujian Medical University Union Hospital, Fuqing City Hospital Affiliated to Fujian Medical University, and Zhangpu County Hospital) between January 2019 and December 2019 were evaluated. Patients who had at least two malignant resected nodules were enrolled in this study. The inclusion criteria were based on the clinical-pathologic criteria [MM criteria and the American College of Chest Physicians (ACCP) modified guidelines] (16). The exclusion criteria of this study were different histologic types or carcinoma in situ, N2/3 involvement, and systemic metastases. Patients without surgical specimens or lost to follow-up were excluded from this study. Clinicopathological data were obtained from a retrospective chart review. Tumor stage was classified according to the 8th edition of the tumor-node-metastasis (TNM) classification for lung cancer. Finally, 53 patients (2.1%) were included. The characteristics of the patients with MPLCs are shown in Table 1.
Table 1
| Factors | Multiple primary lung cancer (n=35) | Favored metastases (n=18) | P value |
|---|---|---|---|
| Age (years), mean | 58.4 | 59.2 | 0.76 |
| FEV1%, mean | 92.8 | 84.9 | 0.03 |
| DLCO (%) | 84.0 | 79.0 | 0.19 |
| Hemoglobin (g/L), mean | 136.1 | 137.4 | 0.73 |
| Neutrophil/lymphocyte, mean | 1.9 | 2.0 | 0.75 |
| Total protein (g/L), mean | 73.8 | 75.4 | 0.82 |
| Albumin (g/L), mean | 44.4 | 45.1 | 0.49 |
| Alkaline phosphatase (IU/L) | 68.4 | 85.3 | 0.003 |
| Creatinine (μmol/L), mean | 63.7 | 61.8 | 0.57 |
| Lactate dehydrogenase (U/L), mean | 190.7 | 186.8 | 0.71 |
| Calcium (mmol/L), mean | 2.4 | 2.4 | 0.74 |
| Cyfra211 (ng/mL) | 2.5 | 2.1 | 0.19 |
| CEA (ng/mL) | 2.4 | 2.8 | 0.60 |
| Tumor number, mean | 2.6 | 2.9 | 0.53 |
| Image diameter difference (cm), mean | 0.9 | 0.8 | 0.66 |
| CT average value difference, mean | 12.2 | 37.5 | 0.82 |
| Max image diameter (cm), mean | 1.8 | 1.6 | 0.55 |
| BMI (kg/m2), mean | 22.6 | 22.4 | 0.85 |
| Gender, n (%) | 0.44 | ||
| Male | 8 (15.1) | 3 (5.6) | |
| Female | 27 (51.0) | 15 (28.3) | |
| Presence of symptoms, n (%) | 0.64 | ||
| No | 29 (54.7) | 15 (28.3) | |
| Yes | 6 (11.3) | 3 (5.6) | |
| Morphological features of image, n (%) | 0.11 | ||
| Same | 1 (1.9) | 3 (5.6) | |
| At least one difference characteristic | 34 (64.2) | 15 (28.3) | |
| Tumor location, n (%) | 0.89 | ||
| Same lobe | 12 (22.6) | 5 (9.4) | |
| Different lobes | 14 (26.4) | 8 (15.1) | |
| Contralateral side | 9 (17.0) | 5 (9.4) | |
| Pathological subtype difference, n (%) | 0.058 | ||
| Consistent | 12 (22.6) | 11 (20.7) | |
| Variation | 23 (43.4) | 7 (13.2) | |
| CT solid component, n (%) | 0.79 | ||
| 0% | 3 (5.6) | 1 (1.9) | |
| <50% | 10 (19.9) | 4 (7.5) | |
| ≥50% | 22 (41.5) | 13 (24.5) | |
| Adjuvant therapy, n (%) | 0.34 | ||
| No | 28 (52.8) | 16 (30.2) | |
| Yes | 7 (13.2) | 2 (3.8) | |
| TKI therapy, n (%) | 0.57 | ||
| No | 34 (64.2) | 17 (32.1) | |
| Yes | 1 (1.9) | 1 (1.9) | |
| Family tumor history, n (%) | 0.56 | ||
| No | 30 (56.6) | 15 (28.3) | |
| Yes | 5 (9.4) | 3 (5.7) | |
| Personal tumor history, n (%) | 0.34 | ||
| No | 28 (52.8) | 16 (30.2) | |
| Yes | 7 (13.2) | 2 (3.8) | |
| Smoke, n (%) | 0.44 | ||
| No | 29 (54.7) | 16 (30.2) | |
| Yes | 6 (11.3) | 2 (3.8) | |
| Surgery methods, n (%) | 0.53 | ||
| Wedge | 2 (3.8) | 1 (1.9) | |
| Segmentectomy | 13 (24.5) | 4 (7.5) | |
| Lobectomy | 20 (37.7) | 13 (24.5) | |
| Stage, n (%) | 0.09 | ||
| IA | 31 (58.5) | 14 (26.4) | |
| IB | 1 (1.9) | 4 (7.5) | |
| IIA | 1 (1.9) | 0 | |
| IIB | 2 (3.8) | 0 | |
| Synchronicity, n (%) | 0.85 | ||
| Synchronous | 28 (52.8) | 14 (26.4) | |
| Metachronous | 7 (13.2) | 4 (7.5) |
BMI, body mass index; CEA, carcinoembryonic antigen; CT, computed tomography; DLCO, diffusing capacity of the lung for carbon oxide; FEV1%, forced expiratory volume in the first second percentage; TKI, tyrosine kinase inhibitor.
A contrast-enhanced thoracic computed tomography (CT) scan was necessary before surgery for all patients. Other routine preoperative assessments comprised chest radiographs, cardiopulmonary function tests, ultrasonography or CT of abdominal and adrenal gland, brain magnetic resonance imaging or CT, and bone scans. The protocol of this retrospective study was approved by the Institutional Review Board of the Fujian Medical University Union Hospital (IRB No. 2021QH029) and the other two hospitals were informed and agreed with the study. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. Individual consent for this retrospective analysis was waived. The enrolled patients had activating mutations in 10 genes, including epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), Kirsten rat sarcoma (KRAS), ROS1, BRAF, neurotrophic tyrosine kinase receptor (NTKR) 1/2/3, MET, RET, ERBB2 and PIK3CA, based on NGS of resected nodules after surgery. A total of 130 nodules were sequenced with a 116-gene panel using NGS.
The following clinical characteristics were collected: date of birth, sex, height, body weight (for determination of body mass index), main past relevant diseases (benign and malignant diseases), smoking status, family tumor history, clinical symptoms (chief complaints mentioned the lung cancer-related symptoms, such as cough, shortness of breath, hemoptysis, chest pain, fever), surgery methods, synchronicity, adjuvant therapy or tyrosine kinase inhibitor (TKI) therapy, diffusing capacity of the lung for carbon oxide (DLCO), and FEV1%.
The laboratory data included measurements of serum proteomic signatures, hemoglobin, total protein, albumin, alkaline phosphatase, creatinine, lactate dehydrogenase, calcium, as well as cyfra211 and carcinoembryonic antigen (CEA) concentrations.
A radiologist reviewed all chest CT scans for the following: proportion of ground-glass opacity (GGO), distribution of solid component (0%, less than 50%, 50% or more of the solid), diameter of nodules in millimeters, location of nodules (same lobe, different lobes, and contralateral side lobe), tumor number, CT average value difference and difference in image features. Pathological files were collected, including the pathological subtype difference and stage (IA, IB, IIA, IIB).
NGS and data analysis
Genomic profiling was conducted at Amoydx (Shanghai, China). DNA was extracted from at least 100 ng of cancer tissue obtained from formalin-fixed, paraffin-embedded (FFPE) tumor samples using a DNA Extraction Kit (Amoy Diagnostics, Xiamen, China) following the manufacturer’s protocols. The coding exons of 116 cancer-related genes and selected introns of 10 genes were captured using a custom hybridization capture panel. The targeted library was analyzed with the Illumina Mid Output V2 Reagent Kit and NextSeq CN500. Sequencing data were processed using the AmoyDx NGS data analysis system (ANDAS Data Analyzer) to identify gene variants. The variant filter criteria requiring absolute mutated allele read counts were set to a valid depth ≥180×, percentage of mutated copies ≥0.5%, mutant copy number ≥2, which allowed for detection of the rarer variants in read percentages of 1%.
We considered patients to have IPM when the tumors had the same or shared mutational genes based on the NGS classification (17,18). Tumors with hotspot gene mutations, such as EGFR L858R and 19del, were adjudicated individually by extended molecular review and synthetic judgment. Patients who exhibited entirely unique mutation profiles in each tumor were classified as having MPLC.
Statistics and survival analysis
Differences in patient characteristics among the MPLC and IPM groups were evaluated using chi-squared tests for categorical variables and one-way analysis of variance tests for continuous variables. For the multivariate analysis, logistic regression was used.
Disease-free survival (DFS) was defined as the interval from surgery to either the first recurrence (local or distant) or the last follow-up date. Overall survival was defined as the interval between the patient’s first radical lung cancer surgery and death. The follow-up period was up to December 31, 2021. Survival was estimated using the Kaplan-Meier method, with differences evaluated by a stratified log-rank test. Multivariable analysis with the Cox proportional hazards model was used to assess the simultaneous effects of prognostic factors on survival, including their interactions. All data were analyzed with SPSS Statistics version 23 (IBM, Chicago, IL, USA). A P value of <0.05 was considered statistically significant.
Patient and public involvement
No patients were involved in setting the research question or the outcome measures, nor were they involved in developing plans for recruitment, design, or implementation of the study. No patients were asked to advise on interpretation or writing up of results.
Results
Patient characteristics
A total of 53 patients with 130 lesions were enrolled in this study. Table 1 summarizes their demographics and clinicopathological characteristics. The cohort comprised 42 female and 11 male patients, eight of whom had a history of smoking. A total of 47 ADCs (36.2%; 3 solid, 6.4%; 6 papillary, 12.8%; 2 micropapillary, 4.3%; 15 lepidic, 17.0%; 28 acinar, 59.6%) and 83 microinvasive adenocarcinomas (MIAs) (63.8%; minor pathological subtypes: 1 papillary, 1.2%; 82 acinar, 98.2%) were included in this study. No significant postoperative complications or perioperative deaths occurred in any of the patients. The median follow-up was 54.6 (interquartile range, 39.6–54.6) months. A total of 16 recurrences were identified during follow-up. No statistically significant correlation was observed between the CT solid component, surgical method, TKI therapy, and adjuvant therapy in terms of the 5-year DFS (log-rank P=0.68, data not shown). One of 4 patients with a 0% CT solid component was determined to have IPM.
Gene panel NGS
Ten-gene and 116-gene panel NGS was used to evaluate the mutation profiles of 130 tumors. The number of tumor mutations of each gene within the 116-gene panel is shown in Figure 1. In our Asian population cohort, EGFR mutations were the most prevalent (58.3%), followed by mutations in BCL2L11 (16.1%), ERBB2 (9.2%), PIK3CA (8.5%) and BRAF (7.7%) within the 116-gene panel. Gene mutations identified using the 10-gene panel are shown in Figure 2. No significant difference in DFS was observed between the mutational statuses of each gene (P=0.66, data not shown).
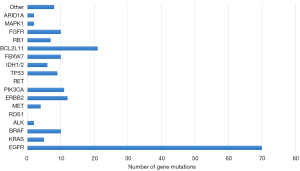
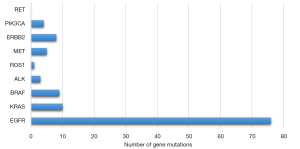
The clonal relatedness of 130 lesions in the 53 cases was identified by mutational profiling. The profiles are shown in Table 2. Patients were also stratified according to their mutation status. Because EGFR is a prevalent gene mutation in Asian populations, we excluded EGFR and divided the patients into two groups (MPLC and IPM groups) according to whether the mutated gene was shared. Based on the 116-gene panel, 18 patients (34%) were classified as having metastases. Based on the 10-gene panel, 5 patients (9.4%) were classified as having metastases. Among the genes with shared mutations, the most common mutation sites were EGFR (n=11, 20.8%) and BCL2L11 (n=11, 20.8%), followed by KRAS (n=1, 1.9%), ERBB2 (n=1, 1.9%), and 1DH2 (n=1, 1.9%) within the 116-gene panel. Significant differences were observed in FEV1% (P=0.03) and alkaline phosphatase (P=0.003) among the MPLC and IPM groups. The multivariate analysis identified FEV1% as an independent factor (P=0.001).
Table 2
| Patient | Lesion | 116-gene mutation | Classification | |
|---|---|---|---|---|
| By 116-gene panel | By 10-gene panel | |||
| 1 | A | EGFR L858R | MPLC | MPLC |
| B | EGFR L747_T751del | |||
| 2 | A | EGFR L858R | IPM | MPLC |
| B | N | |||
| C | EGFR L858R | |||
| D | EGFR L858R | |||
| 3 | A | EGFR L858R | MPLC | MPLC |
| B | EGFR E746_A750del | |||
| 4 | A | EGFR L747S, BRAF T599I | MPLC | MPLC |
| B | N | |||
| 5 | A | N | MPLC | MPLC |
| B | N | |||
| 6 | A | ERBB2 Y772_A775dup | IPM | MPLC |
| B | ERBB2 Y772_A775dup | |||
| C | IDH1 R132H, PIK3CA V344M, TP53 R196* | |||
| 7 | A | N | MPLC | MPLC |
| B | N | |||
| 8 | A | BRAF N581S | MPLC | MPLC |
| B | N | |||
| C | BRAF K601E, KRAS A59T, CTNNB1 T41I | |||
| 9 | A | ERBB2 V773_M774delinsMMAV | MPLC | MPLC |
| B | EGFR L858R | |||
| 10 | A | PIK3CA M1043I, ERBB2 G776delinsVC | IPM | MPLC |
| B | FBXW7 R479Q, EGFR E746_A750del, BCL2L11 c.394+1479_394+4381del | |||
| C | BCL2L11 c.394+1479_394+4381del | |||
| 11 | A | EGFR L858R | MPLC | MPLC |
| B | EGFR L858R | |||
| 12 | A | EGFR L747_T751del, EGFR K754E | MPLC | MPLC |
| B | EGFR L747_P753delinsS | |||
| 13 | A | EGFR L858R | MPLC | MPLC |
| B | MET c.3019_3028+4del | |||
| 14 | A | EGFR L858R, EGFR T790M | MPLC | MPLC |
| B | EGFR G709A, EGFR G719A | |||
| 15 | A | KRAS G13D | MPLC | MPLC |
| B | EGFR L858R, PIK3CA R108H, IDH1 R132H | |||
| 16 | A | N | MPLC | MPLC |
| B | EGFR E746_A750del | |||
| 17 | A | EGFR L858R, BCL2L11 c.394+1479_394+4381del | IPM | MPLC |
| B | EGFR L858R, BCL2L11 c.394+1479_394+4381del | |||
| 18 | A | EGFR L858R, ARID1A R1989* | MPLC | MPLC |
| B | EGFR L858R | |||
| 19 | A | EGFR E746_A750delinsQP, IDH2 R140Q | MPLC | MPLC |
| B | FBXW7 R689W | |||
| 20 | A | EGFR L858R, BCL2L11 c.394+1479_394+4381del | MPLC | MPLC |
| B | BRAF G469V | |||
| 21 | A | EGFR E746_A750delinsQP, BCL2L11 c.394+1479_394+4381del, SF3B1 K700E | IPM | MPLC |
| B | EGFR E709V/G719C, BCL2L11 c.394+1479_394+4381del | |||
| 22 | A | N | MPLC | MPLC |
| B | N | |||
| C | EGFR L858R | |||
| 23 | A | BCL2L11 c.394+1479_394+4381del | IPM | MPLC |
| B | EGFR L858R, BCL2L11 c.394+1479_394+4381del, CREBBP R1446C | |||
| 24 | A | IDH1 R132H, BRAF G469A, TP53 R273H, GNAS R201H, SF3B1 R625H | MPLC | MPLC |
| B | EGFR L858R, EGFR T790M, TERT c.-146C>T, TP53 V173M, PIK3CA G118D | |||
| 25 | A | EGFR L858R, BCL2L11 c.394+1479_394+4381del | IPM | MPLC |
| B | EGFR L858R, BCL2L11 c.394+1479_394+4381del, CTNNB1 D32N | |||
| C | BCL2L11 c.394+1479_394+4381del | |||
| 26 | A | ERBB2 G778_P780dup, RB1 R552*, PIK3CA R108H | MPLC | IPM |
| B | ERBB2 G776delinsVV | |||
| 27 | A | EGFR L747S, EGFR L861Q | IPM | IPM |
| B | EGFR L858R | |||
| 28 | A | N | MPLC | MPLC |
| B | N | |||
| C | EGFR G719A, EGFR L861Q | |||
| 29 | A | EGFR L858R, BCL2L11 c.394+1479_394+4381del | IPM | MPLC |
| B | EGFR E746_T751delinsA, TP53 Q192*, BCL2L11 c.394+1479_394+4381del | |||
| C | EGFR L747_T751del, FBXW7 R658*, BCL2L11 c.394+1479_394+4381del | |||
| 30 | A | EGFR L858R, EGFR E746_A750del | MPLC | MPLC |
| B | FBXW7 R479Q | |||
| 31 | A | FBXW7 R689W, BCL2L11 c.394+1479_394+4381del | IPM | MPLC |
| B | FBXW7 R278*, BRAF K601E, TP53 R273H, TP53 R248Q, ERBB2 G776S, BCL2L11 c.394+1479_394+4381del | |||
| 32 | A | EGFR L858R, TP53 I195N | MPLC | MPLC |
| B | EGFR A767_V769dup, RB1 R320*, NF1 R440* | |||
| 33 | A | EGFR L858R, PIK3CA G118D | MPLC | MPLC |
| B | MAP2K1 Q56P | |||
| 34 | A | EGFR E746_S752delinsV | MPLC | MPLC |
| B | FBXW7 R278*, BRAF G469A | |||
| 35 | A | KRAS G12D | IPM | IPM |
| B | KRAS G12V | |||
| C | N | |||
| D | PIK3R1 R348* | |||
| 36 | A | EGFR E746_A750del, BCL2L11 c.394+1479_394+4381del | IPM | MPLC |
| B | ERBB2 Y772_A775dup, BCL2L11 c.394+1479_394+4381del | |||
| 37 | A | ERBB2 G776delinsVV | MPLC | MPLC |
| B | EGFR N771_H773dup | |||
| C | N | |||
| 38 | A | EGFR E746_A750del, TP53 R213*, ERBB2 R784C, BCL2L11 c.394+1479_394+4381de | IPM | MPLC |
| B | EML4-ALK, BCL2L11 | |||
| 39 | A | PIK3CA R108H, PIK3R1 G376R, APC Q1429*, EGFR R831H, BRAF K601E, BRAF G469E, RB1 R320* | MPLC | MPLC |
| B | EGFR L858R, TP53 R196*/W146* | |||
| 40 | A | EGFR M137R, RB1 R320* | MPLC | MPLC |
| B | KRAS G12V, RB1 R251*, MAPK1 E322K | |||
| C | EGFR A763_Y764insFQEA, FGFR3 R669Q | |||
| 41 | A | N | MPLC | MPLC |
| B | EGFR T790M, EGFR E746_A750del | |||
| C | EGFR L858R, PIK3CA C420R, TP53 M246V | |||
| 42 | A | EGFR L858R, ARID1A R1989*, MAPK1 E322K | MPLC | MPLC |
| B | EGFR L858R | |||
| C | EGFR L861Q | |||
| 43 | A | MET c.3017_3028+5del | MPLC | MPLC |
| B | N | |||
| 44 | A | EGFR L858R | IPM | IPM |
| B | EGFR L858R | |||
| 45 | A | N | MPLC | MPLC |
| B | FBXW7 R465C, EGFR L747_A750delinsP, RB1 R445* | |||
| C | EGFR E709A/G719S, FGFR2 E777K | |||
| 46 | A | EGFR E746_A750del, IDH2 R140Q | IPM | IPM |
| B | RB1 R552*, IDH2 R140Q | |||
| 47 | A | EML4-ALK | MPLC | MPLC |
| B | EGFR L858R | |||
| C | EGFR L858R | |||
| 48 | A | BRAF G469E, BCL2L11 c.394+1479_394+4381del, PIK3CA E707K | IPM | MPLC |
| B | EGFR L858R, BCL2L11 c.394+1479_394+4381del, IDH1 R132H | |||
| C | PIK3CA M1043I, BCL2L11 394+1479_394+4381del, MET c.2888-37_2888-10del | |||
| D | BCL2L11 c.394+1479_394+4381del | |||
| E | BCL2L11 c.394+1479_394+4381del | |||
| F | BCL2L11 c.394+1479_394+4381del | |||
| 49 | A | MET D1228N, ERBB2 Y772_A775dup | MPLC | MPLC |
| B | EGFR L858R, EGFR E746_A750del, ERBB2 A644V, PIK3R1 R348*, PIK3CA E545K/E707K, FBXW7 R278* | |||
| C | EGFR L858R, EGFR T790M | |||
| 50 | A | RAF1 S257L | IPM | MPLC |
| B | EGFR L858R | |||
| C | EGFR L858R | |||
| D | EGFR L858R | |||
| 51 | A | EGFR L858R, EGFR L62R | MPLC | MPLC |
| B | RB1 R320* | |||
| 52 | A | EGFR E746_A750del, BCL2L11 c.394+1479_394+4381del, ARID1A R1722* | IPM | MPLC |
| B | BCL2L11 c.394+1479_394+4381del, RB1 R251* | |||
| 53 | A | EGFR E746_A750del | MPLC | MPLC |
| B | FBXW7 R689W | |||
IPM, intrapulmonary metastases; MPLC, multiple primary lung cancer; N, none of the mutations; NGS, next-generation sequencing.
Survival analysis
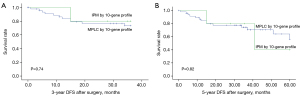
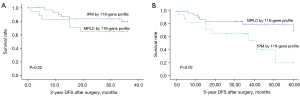
The 3- and 5-year DFS rates were 77.4% and 69.8%, respectively. According to the 10-gene panel, the 3-year DFS was 77.1% and 80% in the MPLC and IPM groups, respectively, and the 5-year DFS rates were 70.8% and 60% in the MPLC and IPM groups, respectively. There was no significant difference found in the 3-year DFS and 5-year DFS between shared or independent mutational profiles (log-rank, P=0.74 and P=0.82, Figure 3). For the 116-gene panel, significance was also not observed for the 3-year DFS (82.9% vs. 66.7%, log-rank P=0.22, Figure 4A). For 5-year DFS, a significant difference was found between the MPLC and IPM groups (80% vs. 60%, log-rank P=0.02, Figure 4B).
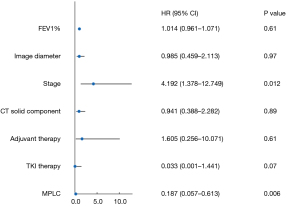
The predetermined covariates were FEV1%, image diameter, stage, CT solid component, adjuvant therapy, and TKI therapy MPLC. The covariates were selected according to multivariate Cox regression analysis. The result is shown in Figure 5. The positive covariates were as follows: stage [hazard ratio (HR) =4.192; 95% confidence interval (CI): 1.378 to 12.749; P=0.01] and MPLC (HR =0.187; 95% CI: 0.057 to 0.613; P=0.006).
Discussion
With the availability of CT, the identification of multiple nodules is becoming more common in clinical practice. The incidence of multiple lung cancers has also increased in recent years. Multiple lung cancers of different histological types are easily diagnosed, but in most cases, multiple lung cancers are challenging to distinguish from metastases (19). MM criteria remain controversial in the diagnosis of MPLC. Recent studies using molecular tools have emphasized the significant limitations of the MM criteria, which remain the most widely used diagnostic criteria (20).
Our cohort showed a significant difference in the 5-year DFS between synchronous and metachronous MPLC (P=0.006, data not shown). Previous studies reported the same findings (21,22). However, other studies had different findings. Goodwin et al. believed that the variation in classification criteria caused significant differences in survival between synchronous and metachronous MPLC (17). We did not find a statistically significant correlation between sex, age, CT solid component, smoking status, surgical method, and adjuvant therapy regarding DFS. A meta-analysis showed the same outcomes (23). We reached the same conclusion as earlier studies (24-27). Stage was significantly associated with 5-year DFS in this cohort (P=0.001, data not shown). Adjuvant chemotherapy and TKI therapy did not show a significant difference in survival. We believe that the low number of stage II patients in this study contributed to this result. Accurate differentiation between MPLC and IPM should support rational treatment strategies and enhance the prognoses of affected patients. Reasonable treatment strategies and improved prognosis depend on the precise distinction between MPLC and IPM. No significant differences in survival curves between different operative techniques (wedge resection, segmentectomy, lobectomy) were found in our study. Our result is similar to what the Japanese JCOG0802 study found in that the results of segmentectomy and lobectomy for solid nodules smaller than 2 cm were comparable (28).
Our innovative discovery was that differences in FEV1% and alkaline phosphatase were observed between the MPLC and PM groups. The findings of multivariate analysis showed that FEV1% was an independent factor. We also tried to analyze whether imaging, including CT average value, image diameter, morphological features of the image, and tumor location, could distinguish MPLC, but the results were negative. This is partly inconsistent with what Araujo-Filho reported (29). This reveals that it is difficult to distinguish MPLC based on clinical and radiographic data.
The prevalence of tumor mutations in our cohort was different from that in other reports. BRAF mutations were found to occur at a higher rate than the reported in previous studies (9,13). The reason for this may be due to population and cohort selection in addition to detecting the less common variants. The same outcomes were observed for other mutations, including EGFR, MET, and ALK (13).
Recently, an increasing number of studies have suggested that gene mutation detection plays a key role in the clonality of tumors (30). Wu et al. described 8 gene mutations (EGFR, KRAS, HER2, BRAF, PIK3CA, ALK, ROS1, and RET) in 72 tumors from 35 patients (13). They found that 5 (14%) patients had clonality and demonstrated that clonality evaluation was possible. Girard et al. assessed clonal relationships using a panel of 101 genes and correlated them to their clinicopathological characteristics (9). They analyzed a total of 42 tumors from 20 patients. Four (18%) cases had discordance compared with the clinicopathologic diagnosis. Our findings are largely consistent with these studies. In our research, classification based on 10-gene profiling contradicted the clinicopathologic diagnosis in 5 (9.4%) of the comparisons, while the discordance was 34% in the 116-gene panel. Our diagnostic criteria referred to the study by Goodwin et al. (17), but there were inconsistencies. Goodwin et al. analyzed gene mutations in Caucasians, in which the most common genetic mutation is KRAS, and determined IPM by using shared gene mutations, including KRAS (17). We think this was inappropriate. Genes with high mutation rates in a population should not be used as evidence of metastasis diagnosis, as the same carcinogenic process may lead to two independent tumors, both exhibiting the same common driver mutation. Therefore, mutations in EGFR (EGFR L858R and 19del) were excluded from our study. However, clonal heterogeneity and tumor evolution models remain subjects of debate. Oncogenic driver mutations are still regarded as relatively stable during disease progression (31). In the process of metastasis, these studies suggested that while tumors may acquire additional mutations, they are very unlikely to lose previously acquired mutations (6,32).
Girard et al. reported using a comprehensive histologic assessment to differentiate MPLC, which only led to a 9% discordance (33). They demonstrated that comprehensive histologic assessment is a cost-effective and quicker method for differentiating between MPLC and IPM. However, such complex and time-consuming histologic assessments are not practical or feasible in routine clinical pathology. Some studies used immunohistochemical scoring to establish mathematical models to distinguish MPLC, which achieved a specific effect. Subjectivity is a massive problem for pathologists manually examining tumor morphology and immunohistochemical staining; these readings are subject to interobserver variability. In our study, 116-gene and 10-gene panel NGS had a 34% and 9.4% discordance, respectively, with MM criteria. In contrast, objectivity and reproducibility are the greatest strength of molecular diagnoses of tumors using NGS. With commercially availability of molecular testing, NGS may be a more economical and reliable option for precise diagnosis.
In our cohort study, compared with those with MPLC, patients determined to have IPM based on 116-gene panel NGS were expected to require a poorer prognosis in terms of the 5-year DFS. There was no significant difference in the 3-year DFS; we suspect this was due to insufficient follow-up time. We used DFS to assess survival because of the high number of patients in the early stages of disease in this study. Classification by 116-gene panel NGS better reflects a difference in survival compared with MM criteria and 10-gene panel NGS. Roepman et al. and Goodwin et al. also reported similar results (12,17). According to the presented data, our study showed that patients determined to have MPLC or IPM have a difference in the 5-year DFS. In contrast, this result is absent if the MM criteria or a 10-gene panel NGS is used. Multivariable Cox regression analysis showed that prognostic factors were associated with tumor stage and MPLC.
Determining the clonality of tumors in clinical practice increasingly relies on NGS especially MM criteria (34), and older mutation profiling techniques have been inconclusive. Currently, cost is an essential factor in analyzing multiple tumors for patients. If NGS analysis can provide more accurate staging as well as prognostic information, then an improved benefit-cost ratio may also be considered. Based on our data showing that they have different stage classifications and prognoses, this study demonstrates that the differentiation of MPLC from IPM is of great interest. Uncertainty persists about the specificity and sensitivity of current diagnostic criteria for making distinctions, which may also impact patient prognosis. Somatic mutations and small panels have a limited role in differentiating MPLC. Large panels may provide more discriminatory information and a more accurate prognosis. With advances in genetic technology, faster, simpler and cheaper genetic analysis offers a reliable alternative to older clinical pathology methods.
To the best of our knowledge, our study is the first attempt to distinguish the diagnosis of multiple pulmonary nodules and the associated prognosis in an Asian population. In our study, we found the way to distinguish between diagnoses. Using this method, the diagnosis results were closely related to the prognosis. This may change clinical practice in the diagnosis and management of multiple pulmonary nodules. There were some limitations in our study. This analysis was based on a relatively small sample size at a single center. Since patients were selected from a specific and limited population within our department, this was a potential source of selection bias. This was a retrospective study which included Asian patients. A prospective study with a diverse population and a large number of patients is required to validate our results.
Conclusions
NGS-based 116-gene panel classification can improve the accuracy of diagnosing MPLC and IPM. The diagnosis of IPM was associated with poor prognosis in Asian population. Researchers should pay more attention to the prognosis and treatment of patients with IPM confirmed by NGS. Meanwhile, NGS can provide additional mutation information for targeted therapy.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-2024-1160/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-2024-1160/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-2024-1160/prf
Funding: This work was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-2024-1160/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The protocol of this retrospective study was approved by the Institutional Review Board of the Fujian Medical University Union Hospital (IRB No. 2021QH029) and the other two hospitals were informed and agreed with the study. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. Individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87-108. [Crossref] [PubMed]
- Johnson BE. Second lung cancers in patients after treatment for an initial lung cancer. J Natl Cancer Inst 1998;90:1335-45. [Crossref] [PubMed]
- van Rens MT, Zanen P, Brutel de La Rivière A, et al. Survival in synchronous vs. single lung cancer: upstaging better reflects prognosis. Chest 2000;118:952-8. [Crossref] [PubMed]
- Martini N, Melamed MR. Multiple primary lung cancers. J Thorac Cardiovasc Surg 1975;70:606-12.
- Detterbeck FC, Marom EM, Arenberg DA, et al. The IASLC Lung Cancer Staging Project: Background Data and Proposals for the Application of TNM Staging Rules to Lung Cancer Presenting as Multiple Nodules with Ground Glass or Lepidic Features or a Pneumonic Type of Involvement in the Forthcoming Eighth Edition of the TNM Classification. J Thorac Oncol 2016;11:666-80.
- Detterbeck FC, Franklin WA, Nicholson AG, et al. The IASLC Lung Cancer Staging Project: Background Data and Proposed Criteria to Distinguish Separate Primary Lung Cancers from Metastatic Foci in Patients with Two Lung Tumors in the Forthcoming Eighth Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:651-65.
- Detterbeck FC, Nicholson AG, Franklin WA, et al. The IASLC Lung Cancer Staging Project: Summary of Proposals for Revisions of the Classification of Lung Cancers with Multiple Pulmonary Sites of Involvement in the Forthcoming Eighth Edition of the TNM Classification. J Thorac Oncol 2016;11:639-50.
- Weinberg BA, Gowen K, Lee TK, et al. Comprehensive Genomic Profiling Aids in Distinguishing Metastatic Recurrence from Second Primary Cancers. Oncologist 2017;22:152-7. [Crossref] [PubMed]
- Girard N, Ostrovnaya I, Lau C, et al. Genomic and mutational profiling to assess clonal relationships between multiple non-small cell lung cancers. Clin Cancer Res 2009;15:5184-90. [Crossref] [PubMed]
- Patel SB, Kadi W, Walts AE, et al. Next-Generation Sequencing: A Novel Approach to Distinguish Multifocal Primary Lung Adenocarcinomas from Intrapulmonary Metastases. J Mol Diagn 2017;19:870-80. [Crossref] [PubMed]
- Saab J, Zia H, Mathew S, et al. Utility of Genomic Analysis in Differentiating Synchronous and Metachronous Lung Adenocarcinomas from Primary Adenocarcinomas with Intrapulmonary Metastasis. Transl Oncol 2017;10:442-9. [Crossref] [PubMed]
- Roepman P, Ten Heuvel A, Scheidel KC, et al. Added Value of 50-Gene Panel Sequencing to Distinguish Multiple Primary Lung Cancers from Pulmonary Metastases: A Systematic Investigation. J Mol Diagn 2018;20:436-45. [Crossref] [PubMed]
- Wu C, Zhao C, Yang Y, et al. High Discrepancy of Driver Mutations in Patients with NSCLC and Synchronous Multiple Lung Ground-Glass Nodules. J Thorac Oncol 2015;10:778-83. [Crossref] [PubMed]
- Takahashi Y, Shien K, Tomida S, et al. Comparative mutational evaluation of multiple lung cancers by multiplex oncogene mutation analysis. Cancer Sci 2018;109:3634-42. [Crossref] [PubMed]
- Mansuet-Lupo A, Barritault M, Alifano M, et al. Proposal for a Combined Histomolecular Algorithm to Distinguish Multiple Primary Adenocarcinomas from Intrapulmonary Metastasis in Patients with Multiple Lung Tumors. J Thorac Oncol 2019;14:844-56. [Crossref] [PubMed]
- Shen KR, Meyers BF, Larner JM, et al. Special treatment issues in lung cancer: ACCP evidence-based clinical practice guidelines (2nd edition). Chest 2007;132:290S-305S.
- Goodwin D, Rathi V, Conron M, et al. Genomic and Clinical Significance of Multiple Primary Lung Cancers as Determined by Next-Generation Sequencing. J Thorac Oncol 2021;16:1166-75. [Crossref] [PubMed]
- Chang JC, Alex D, Bott M, et al. Comprehensive Next-Generation Sequencing Unambiguously Distinguishes Separate Primary Lung Carcinomas From Intrapulmonary Metastases: Comparison with Standard Histopathologic Approach. Clin Cancer Res 2019;25:7113-25. [Crossref] [PubMed]
- Chen C, Huang X, Peng M, et al. Multiple primary lung cancer: a rising challenge. J Thorac Dis 2019;11:S523-36. [Crossref] [PubMed]
- Chen X, Lu J, Wu Y, et al. Genetic features and application value of next generation sequencing in the diagnosis of synchronous multifocal lung adenocarcinoma. Oncol Lett 2020;20:2829-39. [Crossref] [PubMed]
- Sun F, Wang P, Zheng Y, et al. Diagnosis, clinicopathological characteristics and prognosis of pulmonary mucinous adenocarcinoma. Oncol Lett 2018;15:489-94. [Crossref] [PubMed]
- van Rens MT, Eijken EJ, Elbers JR, et al. p53 mutation analysis for definite diagnosis of multiple primary lung carcinoma. Cancer 2002;94:188-96. [Crossref] [PubMed]
- Jiang L, He J, Shi X, et al. Prognosis of synchronous and metachronous multiple primary lung cancers: systematic review and meta-analysis. Lung Cancer 2015;87:303-10. [Crossref] [PubMed]
- Tanvetyanon T, Robinson L, Sommers KE, et al. Relationship between tumor size and survival among patients with resection of multiple synchronous lung cancers. J Thorac Oncol 2010;5:1018-24. [Crossref] [PubMed]
- Dai L, Yang HL, Yan WP, et al. The equivalent efficacy of multiple operations for multiple primary lung cancer and a single operation for single primary lung cancer. J Thorac Dis 2016;8:855-61. [Crossref] [PubMed]
- Yang CY, Yeh YC, Wang LC, et al. Genomic Profiling With Large-Scale Next-Generation Sequencing Panels Distinguishes Separate Primary Lung Adenocarcinomas From Intrapulmonary Metastases. Mod Pathol 2023;36:100047. [Crossref] [PubMed]
- Chang JC, Rekhtman N. Pathologic Assessment and Staging of Multiple Non-Small Cell Lung Carcinomas: A Paradigm Shift with the Emerging Role of Molecular Methods. Mod Pathol 2024;37:100453. [Crossref] [PubMed]
- Saji H, Okada M, Tsuboi M, et al. Segmentectomy versus lobectomy in small-sized peripheral non-small-cell lung cancer (JCOG0802/WJOG4607L): a multicentre, open-label, phase 3, randomised, controlled, non-inferiority trial. Lancet 2022;399:1607-17. [Crossref] [PubMed]
- Araujo-Filho JAB, Chang J, Mayoral M, et al. Are there imaging characteristics that can distinguish separate primary lung carcinomas from intrapulmonary metastases using next-generation sequencing as a gold standard? Lung Cancer 2021;153:158-64. [Crossref] [PubMed]
- Chou TY, Dacic S, Wistuba I, et al. Differentiating Separate Primary Lung Adenocarcinomas From Intrapulmonary Metastases With Emphasis on Pathological and Molecular Considerations: Recommendations From the International Association for the Study of Lung Cancer Pathology Committee. J Thorac Oncol 2025;20:311-30. [Crossref] [PubMed]
- McGranahan N, Swanton C. Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 2017;168:613-28. [Crossref] [PubMed]
- Jesinghaus M, Wolf T, Pfarr N, et al. Distinctive Spatiotemporal Stability of Somatic Mutations in Metastasized Microsatellite-stable Colorectal Cancer. Am J Surg Pathol 2015;39:1140-7. [Crossref] [PubMed]
- Girard N, Deshpande C, Lau C, et al. Comprehensive histologic assessment helps to differentiate multiple lung primary nonsmall cell carcinomas from metastases. Am J Surg Pathol 2009;33:1752-64. [Crossref] [PubMed]
- Klempner SJ, Ou SH, Costa DB, et al. The Clinical Use of Genomic Profiling to Distinguish Intrapulmonary Metastases From Synchronous Primaries in Non-Small-Cell Lung Cancer: A Mini-Review. Clin Lung Cancer 2015;16:334-339.e1. [Crossref] [PubMed]


