Comparison of methods for circulating cell-free DNA isolation using blood from cancer patients: impact on biomarker testing
Introduction
Cancer is among the leading causes of morbidity and mortality worldwide. The incidence of cancer has increased over the years and this trend is expected to rise in the near future (1-3). Recent breakthroughs in the understanding of the biological processes underlying cancer development have led to more effective treatment strategies and targeted therapies have irrevocably changed the treatment of cancer patients. Yet, the use of such therapies requires the biomarker testing in tumor biopsy and obtaining sufficient amount of tumor material for the analysis of somatic mutations in targetable genes is somehow challenging.
Circulating free DNA (cfDNA) released by tumor cells preserves the characteristics of the tissue of origin. As a result, its study permits the genetic characterization of the tumor by a non-invasive procedure and solves the difficulties of conventional biopsy (4-6). Moreover, unlike solid biopsies, in the liquid biopsy circulating tumor DNA (ctDNA) can be quantified over the course of treatment, which can potentially serve to measure tumor response to treatments (7,8). In such scenario, the number of samples to test is expected to increase substantially being the development of high-throughput technologies for cfDNA isolation an important challenge to address. In this way, the emergence of automated systems could improve the reproducibility and robustness of the process and facilitate the implementation of liquid biopsy in the clinical setting.
The aim of this study is to compare the performance two automated systems for cfDNA isolation namely; MagNA Pure Compact (MPC) Nucleic Acid Isolation Kit I (Roche Diagnostics, Penzberg, Germany) and Maxwell® RSC (MR) ccfDNA Plasma Kit (Promega Corporation, Madison, WI, USA) and the later with the QIAamp Circulating Nucleid Acid (QCNA) Kit (QIAgen, Valencia, CA, USA) manual extraction kit, which is one of the most widespread method for cfDNA isolation. Specifically, we compare the cfDNA extraction yield and fragment size of nucleid acids obtained. Finally, we investigate whether the identification of tumor specific mutations could be affected by cfDNA isolation methodologies, by comparing mutant allele fraction (MAF) measurements by digital PCR (dPCR) in 38 cfDNA samples from cancer patients with tumors harboring driver mutations.
Methods
Study population
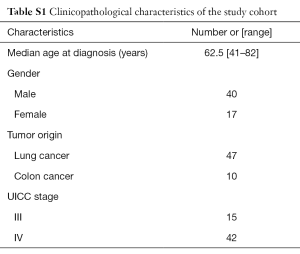
A total of 57 samples were obtained from cancer patients that were prospectively enrolled in the study between February and June 2015 after signing the appropriate informed consent. The study was approved by the Hospital Puerta de Hierro Ethics Committee. The cohort consisted of adult men and women diagnosed with lung cancer or colon cancer, with clinical stage III–IV (Table S1). Information regarding demographics, clinicopathological features, and tumor mutation status was obtained from the clinical and pathology reports.
Full table
Laboratory procedures
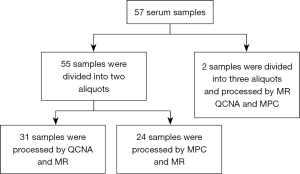
Fifty-seven peripheral whole blood samples were collected in a 4 mL PPT™ tube (Becton Dickinson) containing a gel barrier to separate the plasma after centrifugation. All samples were processed at room temperature within 2 h from the time of blood extraction. Plasma was separated from the cellular fraction by centrifugation at 1,500 g for 10 min at 4 °C. After centrifugation, 55 plasma samples were each divided into two aliquots of 1 mL and two plasma samples were each divided into three aliquots of 1 mL (Figure S1). Aliquots were stored immediately at −80 °C until cfDNA extraction. Hemolyzed samples were discarded for further analysis.
At the time when the cfDNA extraction was performed, samples were thawed in the refrigerator at 4 °C. Once thawed, a new centrifugation was performed at 5,000 g for 20 minutes to ensure removal of impurities in the supernatant.
For cfDNA isolation methodologies comparison purposes, 31 samples were processed by both MR and QCNA, 24 samples were processed by MPC and MR and two plasma samples were aliquoted into three and processed by all three methods (Figure S1).
cfDNA was isolated with a MR instrument (Promega), using the MR ccfDNA Plasma Kit, as specified by the manufacturer and following the “1 mL cell free DNA custom” program, using QCNA Kit (QIAgen), according to manufacturer’s instructions (except when proteinase K incubation was performed overnight) and using the MagNA Pure Compact Nucleic Acid Isolation Kit I on a MPC instrument (Roche Diagnostics).
In all cases, cfDNA was extracted using as starting volume 1 mL of plasma and eluted in 50 µL of the supplied elution buffer.
cfDNA concentration was measured using the Qubit dsDNA HS Assay kit (Invitrogen, Life Technologies, CA, USA) on a Qubit 2.0 Fluorometer (Invitrogen, Life Technologies, CA, USA) according to manufacturer’s instructions.
Microfluidic electrophoresis using the Agilent 2100 Bioanalyzer and High Sensitivity DNA Chips (Agilent technologies Inc., Palo Alto, CA, USA) was performed to assess DNA fragment length for a size range between 50 and 7,000 base pairs (bp) based on manufacturer’s recommended protocol.
cfDNA samples were analyzed by dPCR, using rare mutation assays for the following mutations in EGFR p.T790M (AHRSROS), p.L858R (AHRSRSV) and p.E746_A750delELREA (AHLJ0XO); and mutations p.G12V (AHX1IHY), p.G12D (AH6R5PI) and p.G13D (AHD2BW0) in KRAS, on QuantStudio® 3D Digital PCR System (Applied Biosystems, South San Francisco, CA, USA). For the dPCR, 8 µL of template cfDNA was mixed with 0.5 µL of the above mentioned 40× TaqMan assays and 10 µL of 2× QuantStudio 3D Master Mix, in a 20 µL reaction volume. Subsequently, 15 µL were loaded into QuantStudio 3D Digital PCR 20K chips. The cycling conditions were as follows: initial denaturation at 96 °C for 10 min, followed by 40 cycles at 56 °C for 2 min, and 98 °C for 30 s, a step of 72 °C for 10 min and finally samples were maintained at 22 °C for at least 30 min. Chip fluorescence was read twice. Results were analyzed with QuantStudio® 3D Analysis Suite™ Cloud Software. The automatic call assignments for each data cluster were manually adjusted when needed. The result of the assay is reported as the ratio of mutant DNA molecules relative to the sum of mutant and wild-type (wt) DNA molecules. A wt control DNA was included in every run.
Statistical analysis
Qualitative variables were summarized by their frequency distribution and quantitative variables by their mean and standard deviation (SD) or median and interquartile range. The nonparametric comparison of cfDNA concentration yield by different aliquots of the same plasma sample, using different methodologies, was evaluated using the Wilcoxon signed-rank test for paired data. The nonparametric comparison of cfDNA concentration yield by different clinical situations such as tumor origin (lung cancer vs. colorectal cancer) or tumor staging (stage IV vs. other) was performed using a Mann-Whitney U-test. The association between categorical variables was tested by McNemar’s Test. Null hypothesis was rejected by a type I error minor than 0.05.
Relative quantification of circulating free nucleosome bound DNA fragments was expressed as the ratio between the concentration of a particular fragment and the total concentration of cfDNA in the corresponding sample. Comparison between circulating free nucleosome bound DNA fragments ratios obtained in paired samples using different methodologies was assessed by Wilcoxon signed-rank test for paired data. For this analysis, the level of significance was adjusted to 0.0083 according to the Bonferroni correction for multiple comparisons. The statistical analysis was performed using STATA 9.0 SE.
Results
Comparison of cfDNA yield extracted according to the methodology
The population included in the study was primarily Caucasian (95%) and ranged in age from 41 to 82 years. Among the 57 plasma samples collected, 47 samples corresponded to lung cancer patients and ten samples corresponded to colorectal cancer.
Altogether, we measured the concentration of cfDNA in a total of 116 samples obtained from 57 plasma samples. Overall, the results of quantitation revealed a wide range of cfDNA concentrations in the plasma of cancer patients, ranging between 0.2 and 24 ng/µL.
For comparison purposes, cfDNA was extracted from 33 plasma samples by MR and QCNA. This subset samples were obtained from lung cancer patients, clinical stage III–IV, except for one sample that corresponded to one patient diagnosed with colorectal cancer. According to our results, the median concentration yielded was 1.25 ng/µL for samples extracted by MR methodology and 1.08 ng/µL for samples extracted by QCNA methodology, with no significant differences in yield extraction between methodologies (P=0.775). Interestingly, we observed that the amount of cfDNA obtained from stage IV cancer patients was significantly higher compared to non-metastatic patients. This was true for cfDNA samples isolated using MR and QCNA methodologies (P=0.045 and P=0.0173). Regarding the comparison between MR and MPC methodologies, 26 plasma samples were processed using both methodologies. These plasma samples were obtained from stage IV colorectal cancer patients and lung cancer patients with metastatic disease except for one patient that was IIIA stage. Interestingly, our data showed that MPC extracted significantly less cfDNA than MR (1.154 vs. 2.31; P<0.0001). We also investigate whether tumor location (lung or colon) could influence the cfDNA isolation yield. Our data indicates that there was no significant difference in cfDNA yield according to tumor localization in samples processed by MPC or MR methodology (P=0.9355 and 0.4834 respectively).
Comparison of circulating free nucleosome-bound DNA fragment distribution according to the methodology
Analysis of 44 cfDNA samples obtained from 22 plasma samples extracted with MR and QCNA, and 34 cfDNA samples obtained from 17 plasma samples extracted by MR and MPC methodologies on the Bioanalyzer 2100 was performed in a blinded fashion.
The observed size of the cfDNA fragments was approximately 180 bp (mean 182 bp, range 151–205 bp). In 69 out of 78 cfDNA samples, we also observed a oligonucleosomal DNA ladder pattern as the presence of peaks corresponding to mono-, di- and tri-nucleosomes were clearly visualized in the electrophoretic assay (Figure 1), suggesting an apoptotic cell death. We also detected high-molecular-weight (>10,000 bp) DNA fragments in 54 out of 78 cfDNA samples which could be originate from cells dying via necrosis.
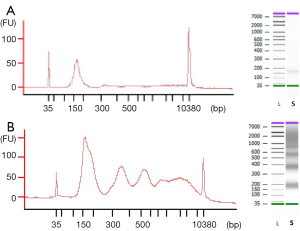
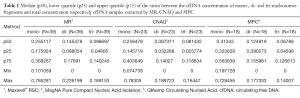
Moreover, the proportion of oligonucleosomal fragments varied depending on the extraction method (Table 1). Remarkably, the proportion of cfDNA fragments ranging in size from 150 to 200 bp extracted by MPC method was significantly higher than by MR method (P=0.0005), whereas the proportion of cfDNA bound to di- and tri-nucleosomes extracted by MPC was significantly lower than by MR (P=0.0024 and P=0.0038).
Full table
Influence of the extraction method on tumor mutation analysis
According to pathology reports, among the 57 cases included, a druggable alteration was found in the corresponding FFPE sample in 26 cases. Of these, 17 cases corresponded to EGFR mutations found in lung cancer tumors and 9 cases corresponded to KRAS mutations found in the colorectal tumors.
Among the 26 patients in which a druggable mutation was found in the corresponding FFPE sample, we were able to detect the identified alteration in 38 paired cfDNA samples isolated from 19 plasma samples, using dPCR. Representative plots for tumor mutation quantification on paired cfDNA samples are displayed in Figure 2. In the remaining cases in which tumor alteration was not detected on the cfDNA, blood samples were extracted during treatment and patients were diagnosed as having a partial response or complete response according to RECIST v1.1 criteria.
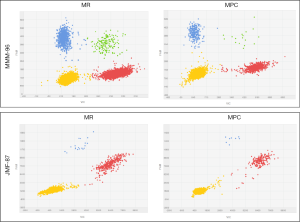
Of the aforementioned 38 cfDNA samples, 30 corresponded to paired cfDNA samples extracted from 15 plasma samples by MR and MPC in parallel. MAF, defined as the proportion of mutant DNA copies relative to the sum of mutant and wt DNA copies, and its confidence interval of all analyzed samples is presented in Table 2. As shown, MAF, on average, was similar between paired cfDNA extracted with MR and MPC, with no significant differences in tumor mutation quantification on cfDNA according to methodology (P=0.8647), suggesting a low impact of extraction method on cfDNA mutation analysis.
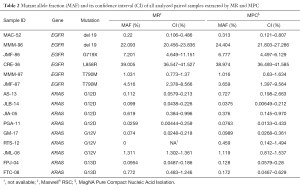
Full table
Finally, eight cfDNA paired samples isolated from four plasma samples by MR and QCNA were analyzed for tumor mutation search and quantification (Table 3). As previously observed MAF was similar between paired cfDNA samples.
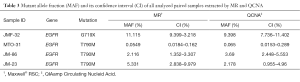
Full table
Discussion
There is a growing evidence supporting that tumor mutation can be effectively detected in blood derived samples such as cfDNA (9-11). Moreover, the relative ease of access and the minimally invasive nature of the sampling make cfDNA an attractive clinical analyte for longitudinal monitoring of cancer disease and response to treatments (7,8). Consequently, it is expected that the number of “liquid” samples that clinical laboratories will manage would be significantly higher compared to FFPE samples. In such scenario, the development of automated systems for cfDNA isolation is an important need to be resolved. Nevertheless, the extraction stage is critical in ensuring reliable results. Remarkably, cfDNA has some peculiarities that should be taken into account as they can profoundly affect the extraction yield and thus downstream applications. Namely, cfDNA concentration is usually very low and cfDNA is highly fragmented with short peak fragment of around 180 bp and its multiples which appeared to correspond to nucleosomal DNA (12,13).
Several studies have compared different extraction methods for the isolation of cfDNA from serum/plasma samples and have indeed demonstrated that the extraction method can considerably affect cfDNA yield (14-17). Similarly, we have observed that MPC and MR extraction methods showed significant differences in the recovery of cfDNA from plasma samples (P<0.0001). In comparing the QCNA kit and the MR, we found that the latter was not superior to the former in terms of cfDNA yield, but it was simpler and more rapid as it is an automated method. To our knowledge, the aforementioned methods comparison has not been reported previously in the literature. In addition, our results illustrate that the kits tested showed significantly different recovery of mono-, di- tri-nucleosomes DNA fragments (Table 1). This observation should be validated in more sized cohort as it has been suggested that low molecular weight cfDNA fractions often harbor genetic aberrations indicative of tumor-derived DNA (17,18).
As previously reported (5), we have found significantly higher cfDNA levels in the plasma of patients with metastatic disease compared to non-metastatic patients. Additionally, we have observed that cfDNA concentration increases with the course of disease being particularly high in dying patients (data not shown). Parallely, it has been described that circulating nucleosomes are in much higher concentration in cancer patients compared to healthy individuals (19-21). Interestingly, it has been reported that among various tumor types, lung cancer is associated with the highest values of circulating nucleosomes (19,20). Similarly, we have observed an oligonucleosomal DNA ladder pattern in most analyzed samples (88%).
The mechanism by which nucleosomes are released into circulation is considered to be apoptotic cell death which is induced by targeted therapies such as tyrosine kinase inhibitors (22,23). This information should be taken into account when tracking mutations on cfDNA for targeted therapies monitoring. If isolation of cfDNA by different procedures can affect the recovery of shorter and longer cfDNA fragments, it is plausible that tumor mutation quantification could be affected by the extraction method. However, we didn’t find significant differences in MAF according to cfDNA extraction methodology. This could in part be due to the small number of samples analyzed and more sized cohort would be of particular interest in order to clarify this issue. Of note, in one sample (RTS-12, Table 2) we could not detect de druggable alteration in the cfDNA isolated by MR but it was detected in the cfDNA extracted by MPC. As mentioned, it has been suggested that low molecular-weight fraction of the cfDNA is enriched in tumor DNA (18,19). According to our results, the recovery of cfDNA fragments ranging in size from 150 to 200 bp by MPC method was significantly higher than by MR method (P=0.0005) although the overall cfDNA extraction yield was significantly lower (P<0.0001). This could explain at least partially the discrepancy observed.
Conclusions
In this paper we demonstrate that there are significant differences in cfDNA extraction yields and high and low molecular-weight cfDNA fractions recovery according to the methodologies evaluated. Larger studies are needed in order to evaluate the impact of such differences on downstream applications such as biomarker testing or tumor mutation tracking for targeted therapies monitoring.
Acknowledgements
The authors wish to thank all the patients that participated in this study.
Funding: This study was supported by Carlos III Institute of Health, Spanish Ministry of Science and Innovation, and European Regional Development Fund (grant number: PI13/01806 and PIE14/00064), A Romero is supported by Joan Rodés fellowship (grant number: JR14/00017) and CP pre-doctoral studies are supported by Jose Luís Castaño Foundation.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the Hospital Puerta de Hierro Ethics Committee (approval number: internal code PI/144-14). Written informed consent was obtained from the patients.
References
- International Agency for Research on Cancer, Cutado MP, Edwards B, et al. Cancer Incidence in Five Continents: Volume IX. Lyon: International Agency for Research on Cancer, 2008.
- Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin 2014;64:9-29. [Crossref] [PubMed]
- American Society of Clinical Oncology. The state of cancer care in America, 2014: a report by the American Society of Clinical Oncology. J Oncol Pract 2014;10:119-42. [Crossref] [PubMed]
- Swarup V, Rajeswari MR. Circulating (cell-free) nucleic acids--a promising, non-invasive tool for early detection of several human diseases. FEBS Lett 2007;581:795-9. [Crossref] [PubMed]
- Leon SA, Shapiro B, Sklaroff DM, et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1977;37:646-50. [PubMed]
- Aung KL, Board RE, Ellison G, et al. Current status and future potential of somatic mutation testing from circulating free DNA in patients with solid tumours. Hugo J 2010;4:11-21. [Crossref] [PubMed]
- Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472-84. [Crossref] [PubMed]
- Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013;368:1199-209. [Crossref] [PubMed]
- He C, Liu M, Zhou C, et al. Detection of epidermal growth factor receptor mutations in plasma by mutant-enriched PCR assay for prediction of the response to gefitinib in patients with non-small-cell lung cancer. Int J Cancer 2009;125:2393-9. [Crossref] [PubMed]
- Kuang Y, Rogers A, Yeap BY, et al. Noninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancer. Clin Cancer Res 2009;15:2630-6. [Crossref] [PubMed]
- Romero A, Acosta-Eyzaguirre D, Sanz J, et al. Identification of E545k mutation in plasma from a PIK3CA wild-type metastatic breast cancer patient by array-based digital polymerase chain reaction: Circulating-free DNA a powerful tool for biomarker testing in advance disease. Transl Res 2015;166:783-7. [Crossref] [PubMed]
- Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001;61:1659-65. [PubMed]
- Suzuki N, Kamataki A, Yamaki J, et al. Characterization of circulating DNA in healthy human plasma. Clin Chim Acta 2008;387:55-8. [Crossref] [PubMed]
- Repiská G, Sedláčková T, Szemes T, et al. Selection of the optimal manual method of cell free fetal DNA isolation from maternal plasma. Clin Chem Lab Med 2013;51:1185-9. [Crossref] [PubMed]
- Fong SL, Zhang JT, Lim CK, et al. Comparison of 7 methods for extracting cell-free DNA from serum samples of colorectal cancer patients. Clin Chem 2009; 55:587-9. [PubMed]
- Holmberg RC, Gindlesperger A, Stokes T, et al. Akonni TruTip(®) and Qiagen(®) methods for extraction of fetal circulating DNA--evaluation by real-time and digital PCR. PLoS One 2013;8:e73068. [Crossref] [PubMed]
- Kuhlmann JD, Schwarzenbach H, Wimberger P, et al. LOH at 6q and 10q in fractionated circulating DNA of ovarian cancer patients is predictive for tumor cell spread and overall survival. BMC Cancer 2012;12:325. [Crossref] [PubMed]
- Schwarzenbach H, Eichelser C, Kropidlowski J, et al. Loss of heterozygosity at tumor suppressor genes detectable on fractionated circulating cell-free tumor DNA as indicator of breast cancer progression. Clin Cancer Res 2012;18:5719-30. [Crossref] [PubMed]
- Holdenrieder S, Stieber P, Förg T, et al. Apoptosis in serum of patients with solid tumours. Anticancer Res 1999;19:2721-4. [PubMed]
- Holdenrieder S, Stieber P, Bodenmüller H, et al. Nucleosomes in serum of patients with benign and malignant diseases. Int J Cancer 2001;95:114-20. [Crossref] [PubMed]
- Holdenrieder S, Stieber P, Bodenmüller H, et al. Circulating nucleosomes in serum. Ann N Y Acad Sci 2001;945:93-102. [Crossref] [PubMed]
- Costa DB, Halmos B, Kumar A, et al. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med 2007;4:1669-79; discussion 1680.
- Faber AC, Li D, Song Y, et al. Differential induction of apoptosis in HER2 and EGFR addicted cancers following PI3K inhibition. Proc Natl Acad Sci U S A 2009;106:19503-8. [Crossref] [PubMed]


