Blood-based tumor biomarkers in lung cancer for detection and treatment
Introduction
Cancer in the 1970s was a biological enigma and the only way forward seemed to utilize combining toxic chemotherapies. However, decades of researches in trying to understand the sentinel secrets of cancer have now brought a paradigm shift in our treatment approach. Especially groundbreaking to this understanding has been the human genome project and the subsequent development of the cancer genome atlas. The advent of targeted therapies in the early 2000s in the form of imatinib and trastuzumab ushered cancer therapy into a new era of personalized medicine. We now know that certain somatic genetic alterations can act as “driver mutations” that can transform cells into malignant entities. Transformed cells rely heavily on these mutations and thus serve as an effective “Achilles heel” to target. For NSCLC in particular, a host of targetable activating mutations have been found including EGFR, ALK, ROS1, KRAS, HER2, MET, RET and FGFR to name a few. The lung cancer genome project has provided invaluable information on genomic-based classification of lung tumors (1). Fifty percent of all NSCLC will be found to have a targetable mutation (2,3). The last decade has seen a robust expansion of novel targeted drugs that deprive these oncogene addicted tumor cells of their vitality. However, despite the impressive initial responses to targeted therapy, the tumors evolve and develop resistance over time. This then requires considerable “ostinato rigore” on the part of patients as well as those treating cancer, to identify new ways to target cancer.
Getting a tissue biopsy has been the mainstay of diagnosis and remains the gold standard in most cases. Along with establishing the histological diagnosis, it is now a standard practice to test for driver mutations by fluorescent in-situ hybridization (FISH) or more recently next-generation sequencing (NGS). However, tissue biopsies are invasive, cumbersome and only allow for a snapshot in time of the ever-evolving tumor biology. It may also miss important tumor characteristics or mutations owing to tumor heterogeneity and small tumors may require multiple attempts to retrieve enough tissue. The ideal test needs to encompass an accurate representation of the tumor biology and heterogeneity. It also needs to be cost efficient, easily collectable, operator independent and have a reasonable turn-around time.
The idea of blood-based biomarkers is not new. Tumor protein biomarkers such as alpha-feto protein (AFP), carcinoembryonic antigen (CEA), cancer antigen 19-9 (CA 19-9) etc. have been utilized for decades to detect disease recurrence, progression, and response to therapy. Although the utility of these tumor markers has been well established in clinical practice, they are not entirely specific and can be elevated in non-malignant conditions. In addition, they do not provide any predictive information on response to therapy, necessitating the need for markers that can furnish elaborate details of tumor biology and guide treatment strategies. Over the past several years, a number of such predictive blood-based biomarkers have entered the scientific research arena. These broadly include circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), tumor-derived exosomes, tumor-educated platelets (TEPs) and miRNAs. Blood based biomarkers are non-invasive and have the potential of assessing real-time tumor response to therapy as well as identifying dynamic resistant clones. As technologies mature, they are becoming faster and more accurate. A summary of available techniques to obtain clinically useful information from tissue and liquid biopsies is depicted in Figure 1.
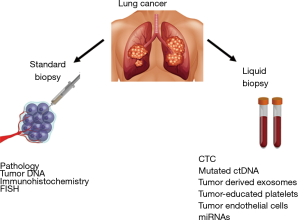
Currently available blood-based biomarkers
CTCs
Viable tumor-derived cells have been identified in peripheral blood from cancer patients and are probably the origin of intractable metastatic disease. Although extremely rare, CTCs represent a potential alternative to invasive biopsies as a source of tumor tissue for the detection, characterization and monitoring of non-hematologic cancers (4-7). The presence of CTCs has been associated with poor outcome in metastatic NSCLC patients as well as in other tumors (8,9). Although CTCs were first described in 1869, their potential utility in guiding treatment of various malignancies has not been fully realized (10).
The technology to obtain these cells is an evolving field of research and is challenged by the ability to isolate CTCs in a condition that can be utilized for molecular analysis and propagation into CTC derived xenografts. After an initial enrichment step that increases the concentration of CTCs by several log units, CTCs can be positively or negatively enriched on the basis of their biological properties (i.e., expression of protein markers) or on the basis of physical properties (i.e., size, density, deformability, or electric charge) (11). A number of CTC detection methods are currently available or under development, which include bench-top instruments, such as flow cytometers (12), high-definition fluorescence scanning microscopy (13), fiber-optic array scanning technology (FAST) (14), isolation by size of epithelial tumor cells (ISET) (15), and laser scanning cytometers (16); while CTC microdevices utilize miniature structure (17), microfluidic reaction kinetics (4), microchip with immunomagnetics (18) and integrated processes (19). In general, CTC microdevices demonstrate superior sensitivity and better cell recovery (20). One of the most validated, and FDA approved, method of obtaining CTCs is CellSearch. CellSearch utilizes ferroparticles and antibodies directed at various epithelial targets such as EpCAM, and cytokeratins (CK 8, 18, and 19) to identify CTCs. CellSearch, while it is highly reliable, can yield false positive results in some benign and inflammatory conditions including: diabetes mellitus, thyroid disorders, hypercholesterolemia and benign breast conditions (21). Low numbers of CTCs specifically in NSCLC limits the ability to routinely collect samples for analysis through CellSearch and magneto-fluidic systems, possibly due to low EpCAM expression in these cells (22). An alternative method of obtaining CTCs utilizes the malignant cell’s difference in physical size and deformity compared to normal cells (23). Micro-ellipse filters allow smaller malleable cells to filter through leaving behind the larger irregular CTCs (23). In contrast to CellSearch, this method allows the collection of EpCAM negative cells, but is limited by potentially missing smaller CTCs.
In resectable NSCLC, CTCs are detected in 19% to 39% of patients by CellSearch analysis, and in 36% to 50% by the ISET method (24,25). A prospective study evaluating CTCs in 150 patients with a suspicious or a diagnosis of primary lung cancer using the CellSearch system investigated the diagnostic role of CTCs by discriminating lung cancer from non-malignant disease. Although CTC count was numerically higher in lung cancer patients compared to patients with non-malignant disease, the receiver operating characteristic (ROC) curve did not disclose a good discrimination between lung cancer patients and healthy controls (24,26).
In advanced NSCLC, CTC counts are generally higher and studies have shown 32% to 78% positivity by CellSearch and up to 80% by ISET (27,28). Data from different laboratories have repeatedly shown limited consistency of results obtained by the CellSearch and ISET approaches. At least 2 studies directly comparing CellSearch and ISET in the same patient cohort have shown low concordance rates (25). These inconsistences are probably explained by differences in the subpopulations of the CTCs measured by each technique, use of different types of antibodies for CTC enrichment, and inherent differences in the sensitivities of the tests (29). Despite these inconsistencies, several studies have shown correlation between number of CTCs and prognosis of patients with advanced NSCLC. In a single center prospective study of 101 patients with previously untreated advanced NSCLC, Krebs and colleagues observed that the number of CTCs in 7.5 mL of blood was higher in patients with stage IV NSCLC compared with patients with stage IIIB or IIIA disease. The progression-free survival (PFS) was 6.8 vs. 2.4 months and overall survival (OS) was 8.1 vs. 4.3 months for patients with fewer than 5 CTCs compared with 5 or more CTCs before chemotherapy, respectively. In multivariate analysis, CTC number was the strongest predictor of OS, and the point estimate of the HR was increased with incorporation of a second CTC sample that was taken after one cycle of chemotherapy (27). Similarly, another study with 40 patients that looked at correlation of CTC number with radiographic tumor response showed that higher baseline CTC counts were associated with response to treatment by Response Evaluation Criteria in Solid Tumors (RECIST) and decreased CTC counts upon treatment were associated with FDG-PET and RECIST response and longer PFS (30). In contrast to these results, a retrospective study of 71 patients with advanced NSCLC showed that the number of CTCs had a weak correlation with the tumor standardized uptake value (SUV) on FDG PET scan and not correlated with tumor diameter. For a given partial volume corrected SUVmax or tumor diameter there was a wide range of detected CTCs in circulation for both early and late stage disease (31). These results suggest that the number of CTCs can potentially serve as prognostic biomarker in advanced NSCLC, although its association with tumor burden is no quite clear.
The utility of CTCs in predicting recurrence after surgery in early stage NSCLC was shown in a study of 56 patients where 51.8% of the patients did have CTCs prior to surgery. The mean number of CTCs was 3.16 per 10 mL preoperatively and 0.66 one month after the surgery. The presence of CTCs after surgery was significantly associated with early recurrence and a shorter disease free survival (DFS). In multivariate analysis, CTC presence after surgery and nodal status were independent prognostic factors for DFS (32). Although these results need further validation in larger patient cohorts, they are certainly encouraging and may eventually identify a subset of patients who can be safely observed without adjuvant chemotherapy.
Another study presented at AACR multidisciplinary thoracic cancers symposium in 2017 reported CTC count of 48 patients with NSCLC collected before, during, and after concurrent chemoradiation. Of 48 patients, 15 had disease recurrence, all of whom had no detectable CTCs following treatment, but the count rose in subsequent tests. In 10 cases, this increase was detectable an average of 6 months before radiographic evidence of recurrence.
Finally, CTC derived xenografts is an exciting area of research in recent years. Patient derived xenograft (PDX) models have gained popularity in cancer research and are used for preclinical drug evaluation, biomarker identification, biologic studies, and personalized medicine strategies. Recently, CTCs have been used to generate PDX experimental models of breast and prostate cancer (33). Advantages of this method over conventional PDX models include independence from surgical sample collection and generating experimental models at various disease stages. CTC derived xenografts have the potential to more accurately reflect the biology of a patient’s cancer rather than tumor cell lines that have been propagated for prolonged periods of time in tissue culture as these are usually homogenous and do not accurately reflect the genetic diversity and constantly adapting microenvironment of cancer. CTC derived xenografts on the other hand can provide a window through which we can constantly study a dynamic evolution of cancer. The primary limitation of this method is the negative selection method used for CTC enrichment. Despite this limitation, the generation of PDX models from CTCs provides a novel experimental model that can be utilized for new drug development. A recent study from small cell lung cancer (SCLC) patients showed the response of CTC derived xenografts to platinum correlated with patient response to platinum and accurately identified chemosensitive and chemoresistant patients. SCLC’s early dissemination provides the ability to obtain CTCs which are currently being studied to evaluate the tumors genome, chemo-sensitivity and chemo-resistance (34). The liquid biopsies allow for the possibility to monitor tumor DNA changes leading to chemoresistance. Further evaluation of CTC derived xenografts in immunocompromised mice have preserved morphologic and genetic characteristics and correlated with platinum response (34).
In summary, CTCs expand our ability to isolate tumor cells from the peripheral blood without the need for invasive and expensive biopsy procedures. In addition, CTCs may serve as the only tool in guiding treatment in patients where invasive biopsy is not feasible because of the location of the tumor or patient’s comorbidities. Along with initial evaluation of tumors, CTCs allow monitoring of the longitudinal evolution of a tumor at the molecular level thereby guiding diagnosis, prognosis, and treatment decisions. Nevertheless, molecular studies of CTCs are not without limitations. One of the major challenges with CTCs is to obtain tumor cells in adequate number and optimum condition that can be utilized for further evaluation. Furthermore, multiple blood samples may require to be collected at different times to get a sample that allows capturing of sufficient number of CTCs. Finally, the technology to assess molecular characteristics of CTCs is still evolving and needs standardization before it can be utilized in routine clinical practice. The utility of CTCs in tailoring targeted therapy in tumors harboring actionable mutations is discussed subsequent sections.
ctDNA
CtDNA, although first identified in 1977 has gained more relevance only recently in the last decade as gene sequencing technologies became faster, cheaper and more accurate. CtDNA is a single strand (35) or double stranded DNA, shed by either living, dying tumor or CTCs into the blood (36-38). Isolating and sequencing tumor DNA among a milieu of normal DNA was a slow and laborious process using the traditional Sanger based sequencing methods. However new BEAMing (beads, emulsion, amplification, magnetics) technology and CAPP-seq (cancer personalized profiling by deep sequencing) have changed the landscape of ctDNA. These techniques amplify target DNA by using already known tag sequencing primers. CAPP-seq can even quantify ctDNA and can identify mutations in 100% of stage II-IV and 50% of stage I NSCLC patients (39,40). One of the limitations of this study is that the target has to be known in order to identify it. Untargeted ctDNA approaches like whole-exome and whole-genome sequencing give a more comprehensive picture.
Exosomes
Exosomes are vesicles of endocytic origin with a diameter of 40−100 nm that transfer information (including proteins, DNA, and RNA) to the target cells through fusion with the plasma membrane, receptor-ligand interaction with the cell or endocytosis by phagocytic mechanism (41). Exosomes are of pivotal importance in tumor biology including local growth, metastasis, and drug resistance by transferring oncogenic proteins and nucleic acids to the tumor cells. Therefore, exosomes and their content could potentially serve as valuable biomarkers in diagnosis, prognosis, and prediction of response to treatment (42). A number of techniques have been used and are under development for exosome isolation, such as MACS (Magnetic Activated Cell Sorting), immune-mediated isolation, sucrose gradient method, and ultra-centrifugation. Once isolated, Western Blot, quantitative RT-PCR, nucleic acid sequencing, ELISA, and other commercially available kits can be utilized to characterize the RNA and protein contents of the exosomes (43). The levels of circulating exosomes and their contents (RNA and miRNAs) have been studied to elucidate their potential role in guiding diagnosis and predicting prognosis of patients with adenocarcinoma of the lung (44). Additionally, the similarity found between the circulating exosomal miRNA and the tumor-derived miRNA patterns led the authors to suggest that circulating exosomal miRNA might be useful as a screening test for lung adenocarcinoma. Another study has reported a model based on microRNAs derived from circulating exosomes capable to discriminate between lung adenocarcinoma and granuloma (45). Finally, the RNA protected in vesicles as the exosomal RNA appears to be a promising source for the analysis of EML4-ALK fusion transcript (46). None of these techniques are currently in a state to be utilized in clinical practice; however, the preliminary results are promising.
Tumor educated platelets (TEP)
Platelets, traditionally known for their role in hemostasis, play salient role in the systemic and local responses to tumor growth (47,48). Transfer of tumor-associated biomolecules to the platelets via confrontation with the tumor cells results in sequestration of such biomolecules giving rise to TEP (49,50). In addition, platelets undergo specific splicing of pre-mRNAs in response to activation of platelet surface receptors (51,52), which gives rise to unique mRNA profile that can be potentially utilized for cancer diagnostics (48,53). A study comparing the platelet mRNA profiles of cancer patients with those of healthy subjects indicated that the levels of 20 non-protein coding RNAs were altered in TEPs as compared to platelets from healthy individuals (48). A total of 1,453 out of 5,003 mRNAs were increased (pertaining to hypoxia, differentiation, and immunodeficiency pathways) and 793 out of 5,003 mRNAs were decreased (pertaining to translation, RNA processing, and viral replication) in TEPs as compared to platelet samples of healthy donors. Moreover, TEP mRNA profiles allowed differentiation of patients with KRAS mutant tumors from KRAS wild-type tumors, EGFR mutant tumors, and MET overexpression in NSCLC patients. However, the number of samples analyzed was relatively small along with the risk of algorithm overfitting (48). Nonetheless, this study provides an early signal that TEPs can be potentially utilized as a blood-based biomarker of lung cancer in future.
Circulating RNA and micro RNA (miRNA)
MiRNAs are intracellular non-coding RNA molecules that are 19–22 nucleotides in length. The miRNAs play a key signaling role in a number of tumor types by mediating post-transcriptional silencing (54). The miRNA expression profile of NSCLC or lung cancer cell lines is different from the one found in non-malignant tissues. One study comparing the expression of 15 different miRNAs between squamous cell lung cancer (SCC) and healthy lung tissue showed that 2 miRNAs (let-7e and miR-125a) were downregulated while rest of the 13 miRNAs were upregulated in SCC (55). The differences in the expression profiles of miRNAs between NSCLC and healthy lung tissue can be detected in the fine needle aspiration (FNA) samples and therefore can potentially be utilized for diagnostic purposes (56). However, the need for obtaining tumor tissue for this assessment is associated with the drawbacks of obtaining conventional tissue biopsy. Therefore, there is growing interest in analyzing circulating miRNA as a non-invasive biomarker for NSCLC diagnosis, monitoring response to treatment, and potentially to personalize therapy. Although several studies have been conducted and many others are underway to establish the role of circulating miRNA in diagnosis and management of NSCLC, there have been many inconsistences in the reference controls and the study samples used for miRNA isolation. For instance, one study has shown that a 24-miRNA expression panel could distinguish lung cancer patients from healthy controls and the diagnostic power can be further enhanced by adding factors such as age, sex and smoking status into this model (57). Similarly, another 6-miRNA expression panel has been shown to differentiate NSCLC patients from healthy subjects (58). However, in both these studies the authors utilized U6 as the internal control for the normalization, which may be inaccurate as U6 has been reported to be not a suitable endogenous control for the quantification of cell-free miRNAs (59). Furthermore, two additional studies showed differential expression of other miRNA panels in the plasma of NSCLC patients compared to healthy controls (60,61). However, these studies analyzed only vesicle-associated miRNAs, such as exosomes, thereby missing at least a proportion of vesicle-free miRNAs in the circulation. Finally, it has been shown that a two-miRNA panel (miR-19b-3p and miR-29b-3p) analyzing miRNAs from peripheral blood mononuclear cells (PBMCs) obtained from whole blood can be availed to differentiate NSCLC patients from healthy persons implying that miRNAs obtained from serum may not provide unabridged information on tumor miRNA profile (62). On a whole, the studies suggesting potential role of circulating miRNAs as biomarkers in NSCLC are compelling; further validation of the sample requirements, assays utilized for isolation and quantification of miRNAs, and methods used for data analysis nevertheless need more validation.
Table 1 summarizes the advantages and challenges associated with currently available blood-based biomarkers. Table 2 depicts an overview of the utility of blood-based biomarkers in lung cancer management.
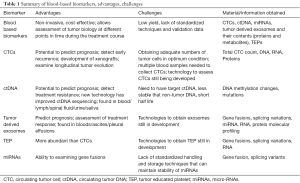
Full table

Full table
Utility of blood based biomarkers in targeted therapy
EGFR
EGFR (epidermal growth factor receptor) is a part of the human EGFR family that includes HER1 (EGFR), HER2/neu, HER3 and HER4. EGFR sensitizing mutations in exons 18-21 lead to propagation of downstream signaling through mTOR and MAP kinase pathways, resulting in increased DNA synthesis and cell proliferation. The most common of these mutations, accounting for almost 90% of all EGFR mutations in NSCLC, results from either an in-frame deletion of Exon 19 (48%) or an L858R substitution on exon 21 (43%). Other less common mutations may also be seen (such as G719X on exon 18, L861Q on exon 21) and both confer sensitivity to small molecule EGFR TKIs.
The role of liquid biopsy in identifying sensitizing EGFR mutations is evolving rapidly. At the moment ctDNA seems to be the most effective means of identifying such mutations by liquid biopsy. An S-ARMS based EGFR mutation detection kit was able to identify the mutation with a sensitivity of 66% and a specificity almost approaching 100% in a sample of 652 patients. Tissue concordance was 94% (73). A COBAS method found similar results in 238 patients reporting 75% sensitivity, 96% specificity and 88% tissue concordance (74). Both studies reported a small population of patients that tested positive by ctDNA but not by tissue largely because the tissue based testing may miss some part of the tumor genomic landscape owing to tumor heterogeneity. However, ctDNA has limited sensitivity and is not a substitute for tissue biopsies yet. It is important to note that ctDNA can yield false negative results in about 15–30% of patients, especially in patients with low volume disease, an indolent lepidic growth pattern, mucinous subtype or occasionally leptomeningeal disease. Improved sequencing technologies such as BEAMing can increase sensitivity to 99% although larger studies are needed to validate their use (75). In 2016 the FDA approved use of COBAS to test for EGFR mutations. The European medical agency suggests using plasma based EGFR testing for patients who can’t get a tissue biopsy. As technology advances and the sensitivity of these tests increases, liquid biopsies may eventually become reasonable alternatives to tissue biopsy.
CTCs can also be used to identify EGFR activating mutations by performing allele specific and deep sequencing NGS techniques. The sensitivity varies between 47–100% in small patient cohorts (76-78). A study suggested using CTCs to monitor for treatment response as gefitinib treatment resulted in decreased CTCs carrying EGFR mutation (79). Isolating CTCs can be difficult, and so can sequencing small CTC populations, therefore its use is currently not recommended. Along similar lines, TEPs may also be found to harbor the EGFR mutations by doing RNA sequencing (48), although further studies are needed to validate its use.
ALK & ROS1
Inversion of chromosome 2p can results in fusion of ALK tyrosine kinase and EML4 protein, leading to constitutive kinase activity and carcinogenesis. This gene rearrangement can be found in 2–7% of patients with NSCLC. Dual tissue IHC and FISH is the best way of identifying the gene rearrangement. However there is inter-operator variability in evaluating FISH, owing to unreliable interpretation of the split signal of the ALK rearrangement. ROS1 has sequence homology to ALK and can be found in 2% of patients with NSCLC.
As with EGFR the potential for liquid biopsies here is immense. However, the routine clinical use of liquid biopsies in detecting ALK and ROS1 mutations is currently limited. A large study of patients showed 94% concordance of liquid biopsies finding actionable mutations in NSCLC using Guardant360 that included the ALK & ROS1 rearrangements. CTCs can also reliably reveal ALK rearrangement via direct FISH analysis (70,80). ROS1 can be detected in CTCs by ISET or FA-FISH (81). CTCs with ROS1 rearrangement increased after stopping crizotinib and correlated with radiological progression of disease. In another study treatment with crizotinib lead to a decreased presence of the ALK positive CTC clone underscoring the potential role for monitoring liquid biopsies for treatment response (82). Other evolving techniques include plasma exosomal RNA sequencing, reported to have a sensitivity of 88% and a specificity of 100%. RT-PCR of TEP RNA can detect the ALK rearrangement and was 65% sensitive and 100% specific (53,83). Crizotinib treatment lead to disappearance of EML-ALK fusion found in TEPs and one patient had re-emergence of the rearrangement 2 months prior to radiological progression, again suggesting it may be helpful for monitoring treatment response.
Uncommon driver mutations
The utility of next generation sequencing of plasma derived ctDNA is not limited to common actionable genetic alterations such as EGFR, ALK, or ROS-1, but can also detect relatively uncommon mutations such as RET rearrangement, MET amplification, and HER-2 insertion. A recent study by Paweletz et al. reported that NGS could identify mutations present in DNA dilutions at ≥0.4% allelic frequency with 100% sensitivity/specificity utilizing a desktop sequencing platform (84).
Utility in managing resistance
Most patients on EGFR TKIs will eventually acquire resistance (85). The most common mechanism of resistance is an acquired exon 20, T790M mutation found in 50–60% of patients who progress on first-line TKI (86). Although tissue biopsy is the mainstay in detecting T790M, there is growing evidence in support of increasing yield by using blood based biomarkers. Tissue biopsies are invasive and not always successful in obtaining viable tissue (78). Oxnard et al. sequenced ctDNA of patients with EGFR-TKI resistance by COBAS and BEAMing techniques and found T790M mutation with a sensitivity of 70% (87). Thirty-one percent of patients with a negative tissue biopsy tested positive by plasma ctDNA. More importantly, progression free survival was similar regardless of means of testing. Thus proceeding with ctDNA testing at the time of EGFR TKI resistance is a reasonable way to identify T790M mutation. However, given the 30% false negative rate seen with ctDNA, tumor biopsy should still follow after a negative plasma test.
Given the need to improve ctDNA based mutation analysis, Huang et al utilized dPCR to detect T790M mutation with increased sensitivity. Presence of T790M detected by this highly sensitive test pre-TKI therapy portended a worse outcome. Ultra-sensitive dd-PCR was able to detect extremely low levels of T790M mutation in 80% of untreated NSCLC patients (86). There is also evidence that serial ctDNA monitoring may pick up T790M mutation, almost a year in advance of clinical progression (67,68,88). Osimertinib is a third generation EGFR TKI active in patients with the T790M resistance mutation (89,90). Twenty-two percent of the patients who progress on osimertinib have C797S resistance mutation. Other means of conferring EGFR TKI resistance include overexpression of c-MET which altogether bypasses the EGFR tyrosine kinase and increases downstream signaling, Her-2 amplification and PI3KCA mutations among others. Approximately 15% of tumors that progress after 1st general TKI, transform in to SCLC which can potentially be detected by identifying SCLC CTCs in the blood. Currently the utility of plasma-based techniques in detecting these other resistance mutations is limited to patients with tumor location that is difficult to biopsy. Moreover, there is still a need to increase the breadth, accuracy and validity of plasma based testing.
Spotlight on blood-based cytokine markers
In addition to advances in genomics based biomarkers, there has been an increasing insight and interest in the effect of lung cancer on proteomics, specifically the cytokine milieu in tumor microenvironment. It is now well known that lung cancer is an immunogenic tumor that makes it susceptible to immune checkpoint inhibitors. However, the immune response to lung cancer is an intricate process with involvement of a number of cytokines and immunologic pathways. Additionally, such process is dynamic and invariably heterogeneous. A “snapshot” of the immunologic/inflammatory milieu of the tumor by means of conventional tissue biopsy is not always sufficient to understand the complex mechanism that renders the tumor resistant to normal immune surveillance. Therefore, theoretically a blood based biomarker that reflects ever changing tumor proteomics and that can be conveniently assessed at different time points during the disease course would be of tremendous value in tailoring cancer directed therapy. Initial attempts at discovering such biomarkers have shown encouraging results, mostly in the context of early detection of lung cancer. For example, Birse et al. compared proteomics of freshly resected lung tumor samples, lung cancer cell lines, and conditioned media collected from tumor cell lines which identified 179 candidate biomarkers. Eight of these markers were selected for further validation study, which showed elevated levels of TFPI, MDK, OPN, MMP2, TIMP1, CEA, CYFRA 21-1, SCC in the serum of patients with Stage I NSCLC compared with high risk smokers without lung cancer (91). Another study comparing circulating inflammatory markers in patients with NSCLC with patients with COPD having similar smoking history showed that concentrations of thymus and activation-regulated cytokine (C-C motif chemokine ligand 17), Gro-b [C-X-C motif chemokine ligand 2 (CXCL2)], CXCL13, interleukin (IL)-1ra, IL-6, IL-8 (CXCL8), IL-16, IL-17A, macrophage migration inhibitory factor (MIF), granulocyte colony-stimulating factor, platelet-derived growth factor subunit B, MMP-2, MMP-8 and MMP-12 were significantly different in serum from NSCLC and COPD patients. In addition, the interferon-γ/IL-10 ratio was lower in cancer patients compared with COPD patients (92). Moreover, peripheral blood mononuclear cell gene expression of CCL3, IL8 and IL1β was found to be higher in lung cancer patients compared to the same patients after removal of their tumors, while CXCL10 and IL2Rα were essentially unchanged (93). Finally, there have been some signals of cytokine signature being of prognostic value as shown in an analysis of patients selected from National Cancer Institute—Maryland case-control study, where patients with stage I NSCLC with high levels of pro-inflammatory cytokines IL-6 and IL-17A had a significantly adverse survival compared with patients with low levels (94). In summary, cytokines as blood-based biomarkers to facilitate early detection and prognostication of early stage lung cancer is an exciting field of discovery which, nevertheless, needs further development and refinement before being used in clinical practice. Moreover, its application in tailoring therapy, specifically immunotherapy, remains to be determined.
Conclusions
In conclusion, the discovery of liquid biopsies in patients with cancer and the recognition of its clinical utility have led to a wealth of studies analyzing the use of blood for cancer diagnostics, prognostication, personalization of treatment, and treatment monitoring. The development of highly sensitive targeted detection methods, the utility and applicability of liquid biopsies for clinical implementation has accelerated. However, current blood-based biosources under evaluation demonstrate suboptimal sensitivity for cancer diagnostics, in particular in patients with localized disease. The ideal test should be sensitive, specific, safe, cost-effective, easily reproducible, and not limited to large academic centers. A prime example of such a test is peripheral blood BCR/ABL transcript level in chronic myeloid leukemia. In spite of significant progress in the arena of blood-based biomarkers, we have not been successful in developing such an ideal test in solid organ malignancies yet. At present, the principal role of blood-based biomarkers in lung cancer is in tailoring targeted therapy in patients with actionable mutations. Its role in selecting chemotherapy remains unclear largely because of the lack of the availability of tissue based biomarker of chemotherapy sensitivity or resistance. The value of liquid biopsy in immunotherapy still remains to be determined. Moreover, most of the biomarkers exploring clinical trials still rely on tissue samples rather than blood samples, primarily because of variability in the methods and accuracies of collecting and analyzing techniques of blood based biomarkers. Nevertheless, currently available blood-based tests on ctDNA and CTCs certainly offer a reasonable surrogate of tissue biopsy in select patient population and appear promising in providing a new dimension to personalized cancer care.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Clinical Lung Cancer Genome Project (CLCGP) Network Genomic Medicine (NGM). A genomics-based classification of human lung tumors. Sci Transl Med 2013;5:209ra153. [PubMed]
- Barlesi F, Mazieres J, Merlio JP, et al. Routine molecular profiling of patients with advanced non-small-cell lung cancer: results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT). Lancet 2016;387:1415-26. [Crossref] [PubMed]
- Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311:1998-2006. [Crossref] [PubMed]
- Nagrath S, Sequist LV, Maheswaran S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007;450:1235-9. [Crossref] [PubMed]
- Mehlen P, Puisieux A. Metastasis: a question of life or death. Nat Rev Cancer 2006;6:449-58. [Crossref] [PubMed]
- Mocellin S, Hoon D, Ambrosi A, et al. The prognostic value of circulating tumor cells in patients with melanoma: a systematic review and meta-analysis. Clin Cancer Res 2006;12:4605-13. [Crossref] [PubMed]
- Pantel K, Alix-Panabieres C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med 2010;16:398-406. [Crossref] [PubMed]
- Rolfo C, Castiglia M, Hong D, et al. Liquid biopsies in lung cancer: the new ambrosia of researchers. Biochim Biophys Acta 2014;1846:539-46. [PubMed]
- Pérez-Callejo D, Romero A, Provencio M, et al. Liquid biopsy based biomarkers in non-small cell lung cancer for diagnosis and treatment monitoring. Transl Lung Cancer Res 2016;5:455-65. [Crossref] [PubMed]
- Ashworth TR. A case of cancer in which cells similar to those in the tumors were seen in the blood after death. Aust Med J 1869;14:146-9.
- Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer 2014;14:623-31. [Crossref] [PubMed]
- Racila E, Euhus D, Weiss AJ, et al. Detection and characterization of carcinoma cells in the blood. Proc Natl Acad Sci U S A 1998;95:4589-94. [Crossref] [PubMed]
- Marrinucci D, Bethel K, Kolatkar A, et al. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Phys Biol 2012;9:016003. [Crossref] [PubMed]
- Krivacic RT, Ladanyi A, Curry DN, et al. A rare-cell detector for cancer. Proc Natl Acad Sci U S A 2004;101:10501-4. [Crossref] [PubMed]
- Pinzani P, Salvadori B, Simi L, et al. Isolation by size of epithelial tumor cells in peripheral blood of patients with breast cancer: correlation with real-time reverse transcriptase-polymerase chain reaction results and feasibility of molecular analysis by laser microdissection. Hum Pathol 2006;37:711-8. [Crossref] [PubMed]
- Rolle A, Gunzel R, Pachmann U, et al. Increase in number of circulating disseminated epithelial cells after surgery for non-small cell lung cancer monitored by MAINTRAC(R) is a predictor for relapse: A preliminary report. World J Surg Oncol 2005;3:18. [Crossref] [PubMed]
- Zheng S, Lin H, Liu JQ, et al. Membrane microfilter device for selective capture, electrolysis and genomic analysis of human circulating tumor cells. J Chromatogr A 2007;1162:154-61. [Crossref] [PubMed]
- Chang CL, Huang W, Jalal SI, et al. Circulating tumor cell detection using a parallel flow micro-aperture chip system. Lab Chip 2015;15:1677-88. [Crossref] [PubMed]
- Adams AA, Okagbare PI, Feng J, et al. Highly efficient circulating tumor cell isolation from whole blood and label-free enumeration using polymer-based microfluidics with an integrated conductivity sensor. J Am Chem Soc 2008;130:8633-41. [Crossref] [PubMed]
- Hong B, Zu Y. Detecting circulating tumor cells: current challenges and new trends. Theranostics 2013;3:377-94. [Crossref] [PubMed]
- Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 2004;10:6897-904. [Crossref] [PubMed]
- Fusi A, Metcalf R, Krebs M, et al. Clinical utility of circulating tumour cell detection in non-small-cell lung cancer. Curr Treat Options Oncol 2013;14:610-22. [Crossref] [PubMed]
- Chen H, Cao B, Sun B, et al. Highly-sensitive capture of circulating tumor cells using micro-ellipse filters. Sci Rep 2017;7:610. [Crossref] [PubMed]
- Tanaka F, Yoneda K, Kondo N, et al. Circulating tumor cell as a diagnostic marker in primary lung cancer. Clin Cancer Res 2009;15:6980-6. [Crossref] [PubMed]
- Wong MP. Circulating tumor cells as lung cancer biomarkers. J Thorac Dis 2012;4:631-4. [PubMed]
- Truini A, Alama A, Dal Bello MG, et al. Clinical Applications of Circulating Tumor Cells in Lung Cancer Patients by CellSearch System. Front Oncol 2014;4:242. [Crossref] [PubMed]
- Krebs MG, Sloane R, Priest L, et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J Clin Oncol 2011;29:1556-63. [Crossref] [PubMed]
- Hofman V, Ilie MI, Long E, et al. Detection of circulating tumor cells as a prognostic factor in patients undergoing radical surgery for non-small-cell lung carcinoma: comparison of the efficacy of the CellSearch Assay and the isolation by size of epithelial tumor cell method. Int J Cancer 2011;129:1651-60. [Crossref] [PubMed]
- Hanssen A, Wagner J, Gorges TM, et al. Characterization of different CTC subpopulations in non-small cell lung cancer. Sci Rep 2016;6:28010. [Crossref] [PubMed]
- Punnoose EA, Atwal S, Liu W, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res 2012;18:2391-401. [Crossref] [PubMed]
- Nair VS, Keu KV, Luttgen MS, et al. An observational study of circulating tumor cells and (18)F-FDG PET uptake in patients with treatment-naive non-small cell lung cancer. PLoS One 2013;8:e67733. [Crossref] [PubMed]
- Bayarri-Lara C, Ortega FG, Cueto Ladron de Guevara A, et al. Circulating Tumor Cells Identify Early Recurrence in Patients with Non-Small Cell Lung Cancer Undergoing Radical Resection. PLoS One 2016;11:e0148659. [Crossref] [PubMed]
- Williams ES, Rodriguez-Bravo V, Chippada-Venkata U, et al. Generation of Prostate Cancer Patient Derived Xenograft Models from Circulating Tumor Cells. J Vis Exp 2015.53182. [PubMed]
- Hodgkinson CL, Morrow CJ, Li Y, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med 2014;20:897-903. [Crossref] [PubMed]
- Sorenson GD, Pribish DM, Valone FH, et al. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol Biomarkers Prev 1994;3:67-71. [PubMed]
- Anker P, Mulcahy H, Chen XQ, et al. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Rev 1999;18:65-73. [Crossref] [PubMed]
- Stroun M, Lyautey J, Lederrey C, et al. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clin Chim Acta 2001;313:139-42. [Crossref] [PubMed]
- van der Vaart M, Pretorius PJ. The origin of circulating free DNA. Clin Chem 2007;53:2215. [Crossref] [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Mittelbrunn M, Sanchez-Madrid F. Intercellular communication: diverse structures for exchange of genetic information. Nat Rev Mol Cell Biol 2012;13:328-35. [PubMed]
- Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl) 2013;91:431-7. [Crossref] [PubMed]
- Zhang X, Yuan X, Shi H, et al. Exosomes in cancer: small particle, big player. J Hematol Oncol 2015;8:83. [Crossref] [PubMed]
- Rabinowits G, Gercel-Taylor C, Day JM, et al. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 2009;10:42-6. [Crossref] [PubMed]
- Cazzoli R, Buttitta F, Di Nicola M, et al. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol 2013;8:1156-62. [Crossref] [PubMed]
- Enderle D, Spiel A, Coticchia CM, et al. Characterization of RNA from Exosomes and Other Extracellular Vesicles Isolated by a Novel Spin Column-Based Method. PLoS One 2015;10:e0136133. [Crossref] [PubMed]
- Leslie M. Cell biology. Beyond clotting: the powers of platelets. Science 2010;328:562-4. [Crossref] [PubMed]
- Best MG, Sol N, Kooi I, et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015;28:666-76. [Crossref] [PubMed]
- Kuznetsov HS, Marsh T, Markens BA, et al. Identification of luminal breast cancers that establish a tumor-supportive macroenvironment defined by proangiogenic platelets and bone marrow-derived cells. Cancer Discov 2012;2:1150-65. [Crossref] [PubMed]
- McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol 2014;16:717-27. [Crossref] [PubMed]
- Schubert S, Weyrich AS, Rowley JW. A tour through the transcriptional landscape of platelets. Blood 2014;124:493-502. [Crossref] [PubMed]
- Rondina MT, Schwertz H, Harris ES, et al. The septic milieu triggers expression of spliced tissue factor mRNA in human platelets. J Thromb Haemost 2011;9:748-58. [Crossref] [PubMed]
- Nilsson RJ, Balaj L, Hulleman E, et al. Blood platelets contain tumor-derived RNA biomarkers. Blood 2011;118:3680-3. [Crossref] [PubMed]
- Hou J, Meng F, Chan LW, et al. Circulating Plasma MicroRNAs As Diagnostic Markers for NSCLC. Front Genet 2016;7:193. [Crossref] [PubMed]
- Raponi M, Dossey L, Jatkoe T, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res 2009;69:5776-83. [Crossref] [PubMed]
- Petriella D, Galetta D, Rubini V, et al. Molecular profiling of thin-prep FNA samples in assisting clinical management of non-small-cell lung cancer. Mol Biotechnol 2013;54:913-9. [Crossref] [PubMed]
- Wozniak MB, Scelo G, Muller DC, et al. Circulating MicroRNAs as Non-Invasive Biomarkers for Early Detection of Non-Small-Cell Lung Cancer. PLoS One 2015;10:e0125026. [Crossref] [PubMed]
- Halvorsen AR, Bjaanaes M, LeBlanc M, et al. A unique set of 6 circulating microRNAs for early detection of non-small cell lung cancer. Oncotarget 2016;7:37250-9. [Crossref] [PubMed]
- Benz F, Roderburg C, Vargas Cardenas D, et al. U6 is unsuitable for normalization of serum miRNA levels in patients with sepsis or liver fibrosis. Exp Mol Med 2013;45:e42. [Crossref] [PubMed]
- Silva J, Garcia V, Zaballos A, et al. Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur Respir J 2011;37:617-23. [Crossref] [PubMed]
- Giallombardo M, Chacártegui Borrás J, Castiglia M, et al. Exosomal miRNA Analysis in Non-small Cell Lung Cancer (NSCLC) Patients' Plasma Through qPCR: A Feasible Liquid Biopsy Tool. J Vis Exp 2016;(111).
- Ma J, Lin Y, Zhan M, et al. Differential miRNA expressions in peripheral blood mononuclear cells for diagnosis of lung cancer. Lab Invest 2015;95:1197-206. [Crossref] [PubMed]
- Alegre E, Fusco JP, Restituto P, et al. Total and mutated EGFR quantification in cell-free DNA from non-small cell lung cancer patients detects tumor heterogeneity and presents prognostic value. Tumour Biol 2016;37:13687-94. [Crossref] [PubMed]
- Guibert N, Ilie M, Long E, et al. KRAS Mutations in Lung Adenocarcinoma: Molecular and Epidemiological Characteristics, Methods for Detection, and Therapeutic Strategy Perspectives. Curr Mol Med 2015;15:418-32. [Crossref] [PubMed]
- Sozzi G, Roz L, Conte D, et al. Plasma DNA quantification in lung cancer computed tomography screening: five-year results of a prospective study. Am J Respir Crit Care Med 2009;179:69-74. [Crossref] [PubMed]
- Wang Z, Chen R, Wang S, et al. Quantification and dynamic monitoring of EGFR T790M in plasma cell-free DNA by digital PCR for prognosis of EGFR-TKI treatment in advanced NSCLC. PLoS One 2014;9:e110780. [Crossref] [PubMed]
- Sorensen BS, Wu L, Wei W, et al. Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non-small cell lung cancer during treatment with erlotinib. Cancer 2014;120:3896-901. [Crossref] [PubMed]
- Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 2014;20:1698-705. [Crossref] [PubMed]
- Hou JM, Krebs M, Ward T, et al. Circulating tumor cells as a window on metastasis biology in lung cancer. Am J Pathol 2011;178:989-96. [Crossref] [PubMed]
- Pailler E, Adam J, Barthelemy A, et al. Detection of circulating tumor cells harboring a unique ALK rearrangement in ALK-positive non-small-cell lung cancer. J Clin Oncol 2013;31:2273-81. [Crossref] [PubMed]
- Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 2008;359:366-77. [Crossref] [PubMed]
- Aieta M, Facchinetti A, De Faveri S, et al. Monitoring and Characterization of Circulating Tumor Cells (CTCs) in a Patient With EML4-ALK-Positive Non-Small Cell Lung Cancer (NSCLC). Clin Lung Cancer 2016;17:e173-7. [Crossref] [PubMed]
- Douillard JY, Ostoros G, Cobo M, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol 2014;9:1345-53. [Crossref] [PubMed]
- Mok T, Wu YL, Lee JS, et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res 2015;21:3196-203. [Crossref] [PubMed]
- Janku F, Angenendt P, Tsimberidou AM, et al. Actionable mutations in plasma cell-free DNA in patients with advanced cancers referred for experimental targeted therapies. Oncotarget 2015;6:12809-21. [Crossref] [PubMed]
- Breitenbuecher F, Hoffarth S, Worm K, et al. Development of a highly sensitive and specific method for detection of circulating tumor cells harboring somatic mutations in non-small-cell lung cancer patients. PLoS One 2014;9:e85350. [Crossref] [PubMed]
- Marchetti A, Del Grammastro M, Felicioni L, et al. Assessment of EGFR mutations in circulating tumor cell preparations from NSCLC patients by next generation sequencing: toward a real-time liquid biopsy for treatment. PLoS One 2014;9:e103883. [Crossref] [PubMed]
- Sundaresan TK, Sequist LV, Heymach JV, et al. Detection of T790M, the Acquired Resistance EGFR Mutation, by Tumor Biopsy versus Noninvasive Blood-Based Analyses. Clin Cancer Res 2016;22:1103-10. [Crossref] [PubMed]
- Kalykaki A, Agelaki S, Kallergi G, et al. Elimination of EGFR-expressing circulating tumor cells in patients with metastatic breast cancer treated with gefitinib. Cancer Chemother Pharmacol 2014;73:685-93. [Crossref] [PubMed]
- Ilie M, Long E, Butori C, et al. ALK-gene rearrangement: a comparative analysis on circulating tumour cells and tumour tissue from patients with lung adenocarcinoma. Ann Oncol 2012;23:2907-13. [Crossref] [PubMed]
- Pailler E, Auger N, Lindsay CR, et al. High level of chromosomal instability in circulating tumor cells of ROS1-rearranged non-small-cell lung cancer. Ann Oncol 2015;26:1408-15. [Crossref] [PubMed]
- Tan CL, Lim TH, Lim T, et al. Concordance of anaplastic lymphoma kinase (ALK) gene rearrangements between circulating tumor cells and tumor in non-small cell lung cancer. Oncotarget 2016;7:23251-62. [Crossref] [PubMed]
- Nilsson RJ, Karachaliou N, Berenguer J, et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget 2016;7:1066-75. [Crossref] [PubMed]
- Paweletz CP, Sacher AG, Raymond CK, et al. Bias-Corrected Targeted Next-Generation Sequencing for Rapid, Multiplexed Detection of Actionable Alterations in Cell-Free DNA from Advanced Lung Cancer Patients. Clin Cancer Res 2016;22:915-22. [Crossref] [PubMed]
- Wheeler DL, Dunn EF, Harari PM. Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat Rev Clin Oncol 2010;7:493-507. [Crossref] [PubMed]
- Arcila ME, Oxnard GR, Nafa K, et al. Rebiopsy of lung cancer patients with acquired resistance to EGFR inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid-based assay. Clin Cancer Res 2011;17:1169-80. [Crossref] [PubMed]
- Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3375-82. [Crossref] [PubMed]
- Watanabe M, Kawaguchi T, Isa S, et al. Ultra-Sensitive Detection of the Pretreatment EGFR T790M Mutation in Non-Small Cell Lung Cancer Patients with an EGFR-Activating Mutation Using Droplet Digital PCR. Clin Cancer Res 2015;21:3552-60. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689-99. [Crossref] [PubMed]
- Birse CE, Lagier RJ, FitzHugh W, et al. Blood-based lung cancer biomarkers identified through proteomic discovery in cancer tissues, cell lines and conditioned medium. Clin Proteomics 2015;12:18. [Crossref] [PubMed]
- Eide HA, Halvorsen AR, Sandhu V, et al. Non-small cell lung cancer is characterised by a distinct inflammatory signature in serum compared with chronic obstructive pulmonary disease. Clin Transl Immunology 2016;5:e109. [Crossref] [PubMed]
- Chang DH, Rutledge JR, Patel AA, et al. The effect of lung cancer on cytokine expression in peripheral blood mononuclear cells. PLoS One 2013;8:e64456. [Crossref] [PubMed]
- Meaney CL, Zingone A, Brown D, et al. Identification of serum inflammatory markers as classifiers of lung cancer mortality for stage I adenocarcinoma. Oncotarget 2017;8:40946-57. [PubMed]


