ROR1 as a novel therapeutic target for EGFR-mutant non-small-cell lung cancer patients with the EGFR T790M mutation
Introduction
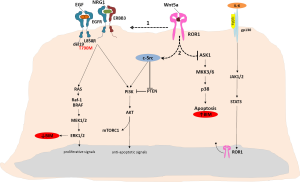
Erlotinib, gefitinib and the second-generation, irreversible epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI), afatinib, have offered a therapeutic alternative for patients with metastatic EGFR positive lung cancer that has proven its superiority over standard platinum-based chemotherapy but without effect on overall survival (1-3). In a large prospective study of EGFR-mutant patients treated with erlotinib, similar overall survival was observed in the first- and second-line settings (4). In the randomized phase III EURTAC trial comparing erlotinib versus chemotherapy in advanced EGFR-mutant patients with non-small-cell lung cancer (NSCLC), the interim analysis confirmed that EGFR mutations were a predictive biomarker of longer progression-free survival (PFS) to erlotinib [hazard ratio (HR) =0.37; P<0.001] (1). However, response was limited to 58% of patients in the erlotinib group and no impact on overall survival was observed (1). Further molecular analyses of 95 patients included in the trial identified the presence of the pretreatment somatic T790M mutation in 65.26% of patients, with a clear impact on PFS to erlotinib (5). In the same group of patients, high BIM mRNA expression was a marker of longer PFS and overall survival (5). Molecular cross-talk between EGFR and other signaling pathways creates alternative means of tumor cell proliferation and promotes resistance to single-agent erlotinib therapy in NSCLC driven by EGFR mutations. Thyroid transcription factor-1 (TTF-1) is a reliable lineage marker for terminal respiratory unit cells as well as for ‘‘terminal respiratory unit-type’’ adenocarcinomas with distinct clinicopathologic and genetic features, including a significant association with EGFR mutations (6). Indeed, TTF-1 has been identified as a lineage-survival oncogene in lung adenocarcinoma and induces expression of the receptor tyrosine kinase (RTK) like orphan receptor 1 (ROR1) (6). ROR1 is a member of the RTK family of orphan receptors related to muscle specific kinase and Trk neurotrophin receptors (7). ROR1 participates in signal transduction, cell-cell interaction, regulation of cell proliferation, differentiation, cell metabolism and survival (7). ROR proteins have emerged as central regulators of Wnt signaling and prior studies have demonstrated that ROR1 serves as a receptor for Wnt5a (Figure 1) (8). Signal transducer and activator of transcription 3 (STAT3) induces both Wnt5a and ROR1 expression in CLL cells (9). Li and colleagues found that interleukin 6 (IL-6) induces STAT3 phosphorylation and upregulates ROR1 protein levels in a time- and dose-dependent manner by activating ROR1 promoter activity (9). ROR1 sustains a favorable balance between prosurvival PI3K-AKT and pro-apoptotic p38 signaling, in part through ROR1 kinase-dependent c-Src activation, as well as kinase-activity independent sustainment of the EGFR-ERBB3 association, ERBB3 phosphorylation, and consequential PI3K activation (Figure 1) (6). ROR1 knockdown effectively inhibited the growth of the gefitinib- or erlotinib-resistant NCI-H1975 lung adenocarcinoma cell line (harboring EGFR L858R and T790M mutations), identifying ROR1 as an “Achilles’ heel” in lung adenocarcinoma that warrants future development of rational synthetic lethality approaches (6).
We examined ROR1 mRNA expression in pretreatment tumor samples from 45 patients included in the EURTAC trial (clinicaltrials.gov NCT00446225), in order to assess its potential role as a predictive biomarker of PFS and overall survival. Twenty-seven patients (60%) harbored pretreatment concomitant EGFR T790M mutations.
Methods
Patients
The EURTAC study enrolled 173 patients with EGFR mutations who were randomized to receive erlotinib or standard intravenous chemotherapy with cisplatin or carboplatin plus docetaxel or gemcitabine (1). We assessed ROR1 mRNA expression in 45 of these patients. The EURTAC was approved by the institutional review board of each participating center, and written informed consent was obtained from all patients.
Molecular analyses
All specimens were formalin-fixed, paraffin-embedded, pretreatment tumor tissues. They were stained with haematoxilin/eosin and evaluated by an expert pathologist. Microdissection was subsequently performed, as previously described (10). All analyses were carried out centrally at the Pangaea Biotech Oncology Laboratory, an ISO 15189-certified laboratory located in the Quiron Dexeus University Hospital (Barcelona, Spain).
ROR1 mRNA expression
Gene expression analysis of ROR1 was performed on RNA isolated from the tumor tissue specimens. RNA extraction, retrotranscription analysis and real-time PCR were performed as previously described (10), and gene expression was examined by qPCR using β-actin as housekeeping gene. Primers and probe for gene expression analysis of β-actin and ROR1 were designed according to their Ref Seq in https://www.ncbi.nlm.nih.gov/sites/entrez?db/gene. Gene expression of β-actin was analyzed using the following primers and probe: forward 5' TGAGCGCGGCTACAGCTT 3'; Reverse 5' TCCTTAATGTCACGCACGATTT 3' and probe 6FAM 5' ACCACCACGGCCGAGCGG 3' TAMRA. Gene expression of ROR1 was analyzed using the following primers and probes: forward 5' CAAGTCCAGGATACTCAGATGAGTATG 3'; Reverse 5' TGCACATGCAATCCCTCTGTA 3' and probe6FAM 5' AGAAGATGGATTCTGTCAGC 3' MGB.
Gene mutations
For assessment of the pretreatment somatic EGFR T790M mutation, cancer cells were microdissected directly into 15 µL of PCR buffer (Ecogen, Barcelona, Spain) plus proteinase K (40 µg/mL) and incubated overnight at 60 °C. Proteinase was inactivated at 95 °C for 10 minutes, and the amount of DNA present in the cell extract was quantified in a Nanodrop and diluted to 5 ng of DNA/µL. Two extracts were obtained from every tumor, from one or more separate tumor areas (10). Cell extracts were directly analyzed by TaqMan in the presence of a peptide-nucleic acid (PNA) designed to inhibit amplification of the wild-type (wt) allele, as previously described (10). In addition, all cell extracts were also assayed in absence of PNA to confirm the presence of DNA. Every tumor sample was run in octuplicate and DNA from the DLD1 cell line at 5 ng/µL was used as a negative control, and DNA from the H1975 NSCLC cell line at 50 pg/µL as a mutated, positive control. The H1975 cell line is derived from an EGFR TKI-naïve patient and harbors both the T790M and L858R mutation as well as mutant TP53 (R273H) [Catalog of Somatic Mutations in Cancer (COSMIC); http://www.sanger.ac.uk/cosmic] (11,12). A tumor was considered positive if at least 1 of the 8 octuplicates showed amplification of the T790M allele in the presence of PNA. Previous validation experiments had demonstrated that our methodology consistently detected the T790M allele in a ratio of 1:5,000 to wild-type alleles.
Statistical analyses
According to the updated results of the EURTAC trial, on 24 January 2013, 135 PFS events had occurred; Results reported here are based on data analyses from that cut-off date (5). For the overall survival analysis, patients were not censored at crossover, while all patients were censored at crossover for the analysis of PFS. PFS and overall survival were estimated by means of the Kaplan-Meier method and compared with a non-parametric log-rank test. A multivariate Cox proportional hazard model was applied with treatment and potential risk factors as covariates, obtaining HRs and their 95% confidence intervals (CIs). Response rates were compared with the χ2 test, or Fisher exact test, as required. The U Mann-Whitney test and the Kruskal-Wallis test were used to test associations between genotypes and clinical characteristics. Each analysis was performed with the use of a two-sided 5% significance level and a 95% CI. The statistical analyses were performed using SAS version 9.2, SPSS version 17.0, or S-PLUS version 6.1. The EURTAC study is registered with ClinicalTrials.gov, number NCT00446225.
Results
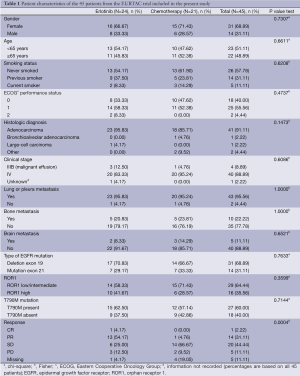
Table 1 shows patient characteristics for the 45 patients included in this sub-analysis of patients from the EURTAC trial (1). The pretreatment somatic EGFR T790M mutation was detected in 27 (60%) patients. The analysis of non-tumor tissue ruled out the presence of germline mutations. Among the 45 patients in whom ROR1 mRNA was successfully examined, ROR1 expression was low (<4) or intermediate (4-6.36) in 29 (64.44%) and high (≥6.36) in 16 (35.56%). There was no statistically significant association between T790M mutation status and ROR1 expression levels.
Full table
Progression-free survival (PFS)
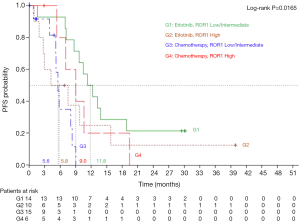
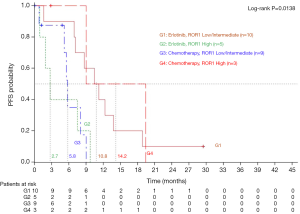
On January 24, 2013, median PFS for all 173 patients in the EURTAC study was 10.4 months for patients treated with erlotinib and 5.1 months for those treated with chemotherapy (HR =0.33; 95% CI, 0.23-0.49; P<0.0001) (5). PFS to erlotinib was 11.8 months (95% CI, 7.1-18.8) for those with low/intermediate ROR1 levels, compared to 5.8 months (95% CI, 0.9-15.8) for those with high ROR1 levels, while among patients treated with chemotherapy PFS was 5.6 months (95% CI, 3.9-8.3) for those with low/intermediate ROR1 levels and 9 months (95% CI, 5.3-19.4) for those with high ROR1 levels (P=0.0165) (Figure 2). Among the 45 patients analyzed for ROR1 mRNA expression, 27 patients had the concomitant pretreatment T790M mutation. In these cases, where T790M was present, PFS to erlotinib was 10.8 months (95% CI, 1.6-13.8) for those with low/intermediate ROR1 levels, compared to 2.7 months (95% CI, 0.9-9.7) for those with high ROR1 levels, while among patients treated with chemotherapy PFS was 5.8 months (95% CI, 0.8-9.0) for those with low/intermediate ROR1 levels, compared to 14.2 months (95% CI, 9.0-19.4) for those with high ROR1 levels (P=0.0138) (Figure 3).
Survival
Overall survival for all 173 patients included in the EURTAC study did not differ between treatment groups (1). On January 24, 2013, with a median follow-up of 40.7 months for the erlotinib group and 35.4 months for the chemotherapy group, median survival was 22.9 months in the erlotinib group and 22.1 months in the chemotherapy group (5). The majority of patients had crossed over at the time of progression. Furthermore, overall survival did not differ among the 45 patients according to ROR1 mRNA levels (Figure 4).
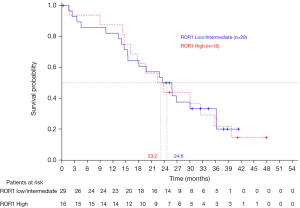
Response
On April 11 2012, overall response rates in the EURTAC study were 65.1% in the erlotinib group and 16.1% in the chemotherapy group (P<0.0001) (5). For the 45 patients with ROR1 mRNA expression, erlotinib-treated patients with low/intermediate ROR1 levels attained an overall response of 71.4%, compared to 40% for those with high ROR1 levels (P=0.058). Among chemotherapy-treated patients, overall response was 6.7% for the 15 patients with low/intermediate ROR1 levels, while the 6 patients with high ROR1 levels did not respond (P=0.361) (Tables 2,3).
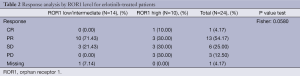
Full table
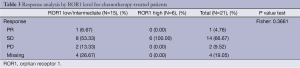
Full table
Discussion
Our study identifies ROR1 as a novel clinical biomarker and therapeutic target that may be used to optimize the outcome of patients with EGFR-mutant NSCLC treated with EGFR TKIs.
ROR1, an evolutionarily conserved type-I membrane protein, is expressed by a variety of different cancers but not by normal postpartum tissues (13). Zhang and colleagues were able to detect ROR1 expression in lymphomas but also in ovarian, colon, lung, pancreatic and prostate cancers cells (8). Cancer cells that are poorly differentiated with high-grade histology are more likely to express ROR1 than tumors with low-grade histology (14). There is also a significant association between expression of ROR1 and activation of PI3K-AKT in cancer cell lines or primary tumor tissues (14). Two recent studies have demonstrated the cell lineage association between EGFR mutations and the homeodomain transcription factor TTF-1 (15,16). Functionally, TTF-1 induced ROR-1 is necessary to sustain the EGFR signaling pathway in lung adenocarcinoma cell lines (6). ROR1, through distinct kinase-dependent and -independent mechanisms, sustains a favorable balance between PI3K-AKT-mediated prosurvival signaling and the pro-apoptotic p38 pathway (6). ROR1 represses proapoptotic p38 signaling, binds to EGFR in response to EGF and leads to activation of the PI3K-AKT pathway by promoting the activation of ERBB3 by EGFR and by phosphorylating c-Src (6). ROR1 physically interacts with and phosphorylates c-Src, a critical component of multiple signaling pathways important for cancer development (6). PTEN negative regulation of PI3K-AKT signaling appears to be, at least in part, under the influence of ROR1 expression, a finding consistent with previous reports on PTEN as a c-Src substrate (6,17) (Figure 1). ROR1 repression inhibits lung adenocarcinoma regardless of EGFR status. Knockdown of ROR1 is sufficient to inhibit growth of lung cancer cell lines with multiple mechanisms of acquired resistance, including EGFR T790M, MET amplification and HGF overexpression (18). ROR1 abrogates apoptosis signal-regulating kinase 1 (ASK1), which can lead to abrogation of the proapoptotic protein BIM (6) (Figure 1). We were the first to reveal that BIM mRNA expression is a unique biomarker of both PFS and overall survival and the identification of the signaling pathways leading to abrogation of BIM can be crucial in the design of synthetic lethality to prevent innate and acquired resistance to EGFR TKI monotherapy (5).
In the present study, ROR1 expression has a differential effect on outcome to erlotinib and chemotherapy in EGFR-mutant NSCLC patients. High levels of ROR1 mRNA expression correlated significantly with poor outcome to erlotinib. PFS was 11.8 months for erlotinib-treated patients with low/intermediate ROR1 levels, compared to 5.8 months for those with high ROR1 levels. RR was 40% for the 10 patients with high ROR1 levels, compared to 71.4% for the 14 with low/intermediate levels. ROR1 expression had the opposite effect on chemotherapy treated patients. PFS was 9 months for EGFR mutant patients who received chemotherapy and had high ROR1 expression levels, compared to 5.6 months for those with low/intermediate ROR1 levels. These differences were stronger when the pretreatment T790M mutation was present. We have recently demonstrated the presence of the pretreatment T790M mutation in 65.26% of EGFR-mutant patients (5). In the present study, 27 of 45 (60%) patients had concomitant pretreatment T790M mutation. When T790M was present, PFS to erlotinib was 10.8 months for those with low/intermediate ROR1 levels, compared to 2.7 months for those with high ROR1 levels, indicating that ROR1-directed therapies can enhance the efficacy of treatment. In contrast, high ROR1 expression confers longer PFS of 14.2 months to chemotherapy in the same group of patients.
To the best of our knowledge this is the first study examining the effect of ROR1 expression in EGFR mutant lung adenocarcinoma patients. Besides hematologic malignancies, the role of ROR1 has been further explored in breast cancer cell lines and tumor samples. Zhang et al., found that breast cancer cells expressing ROR1 generally lack expression of hormone receptors and/or HER2/neu (8). In the study of Cui et al., inhibition of ROR1 with a monoclonal antibody (mAb) specific for the extracellular domain of ROR1 reversed epithelial-to-mesenchymal transition (EMT), allowing breast cancer cells to acquire expression of epithelial cytokeratins and tight junction proteins that may tether the cell to a local microenvironment (14). Salt and colleagues have recently demonstrated the changes in the PI3K-AKT pathway signaling that occur following an EMT (19,20). These data are consistent with a rewiring of PI3K pathway signaling in mesenchymal cells such that it is no longer under the control of ERBB3 but of other RTK, like for instance ROR1, and suggest that these kinases should be targeted following an EMT (14,19).
Our findings suggest that ROR1 may be a therapeutic target in EGFR positive lung adenocarcinomas with or without pretreatment or acquired T790M mutation. A major limitation of our study is the small number of patients with sufficient archival tumor tissue for ROR1 mRNA expression. Still, ROR1 remains an attractive target, worthy of validation in large-scale prospective trials for biomarker development and identification of patients most likely to benefit from rational polytherapies designed to overcome resistance to EGFR TKI monotherapy.
Acknowledgements
Work in Dr Rafael Rosell’s laboratory was partially supported by a grant from La Caixa Foundation.
Disclosure: The authors declare no conflict of interest.
References
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [PubMed]
- Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013;31:3327-34. [PubMed]
- Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009;361:958-67. [PubMed]
- Costa C, Molina MA, Drozdowskyj A, et al. The Impact of EGFR T790M Mutations and BIM mRNA Expression on Outcome in Patients with EGFR-Mutant NSCLC Treated with Erlotinib or Chemotherapy in the Randomized Phase III EURTAC Trial. Clin Cancer Res 2014;20:2001-10. [PubMed]
- Yamaguchi T, Yanagisawa K, Sugiyama R, et al. NKX2-1/TITF1/TTF-1-Induced ROR1 is required to sustain EGFR survival signaling in lung adenocarcinoma. Cancer Cell 2012;21:348-61. [PubMed]
- Daneshmanesh AH, Mikaelsson E, Jeddi-Tehrani M, et al. Ror1, a cell surface receptor tyrosine kinase is expressed in chronic lymphocytic leukemia and may serve as a putative target for therapy. Int J Cancer 2008;123:1190-5. [PubMed]
- Zhang S, Chen L, Cui B, et al. ROR1 is expressed in human breast cancer and associated with enhanced tumor-cell growth. PLoS One 2012;7:e31127. [PubMed]
- Li P, Harris D, Liu Z, et al. Stat3 activates the receptor tyrosine kinase like orphan receptor-1 gene in chronic lymphocytic leukemia cells. PLoS One 2010;5:e11859. [PubMed]
- Rosell R, Molina MA, Costa C, et al. Pretreatment EGFR T790M mutation and BRCA1 mRNA expression in erlotinib-treated advanced non-small-cell lung cancer patients with EGFR mutations. Clin Cancer Res 2011;17:1160-8. [PubMed]
- Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005;2:e73. [PubMed]
- Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 2011;39:D945-50. [PubMed]
- Zhang S, Chen L, Wang-Rodriguez J, et al. The onco-embryonic antigen ROR1 is expressed by a variety of human cancers. Am J Pathol 2012;181:1903-10. [PubMed]
- Cui B, Zhang S, Chen L, et al. Targeting ROR1 inhibits epithelial-mesenchymal transition and metastasis. Cancer Res 2013;73:3649-60. [PubMed]
- Takeuchi T, Tomida S, Yatabe Y, et al. Expression profile-defined classification of lung adenocarcinoma shows close relationship with underlying major genetic changes and clinicopathologic behaviors. J Clin Oncol 2006;24:1679-88. [PubMed]
- Yatabe Y, Kosaka T, Takahashi T, et al. EGFR mutation is specific for terminal respiratory unit type adenocarcinoma. Am J Surg Pathol 2005;29:633-9. [PubMed]
- Lu Y, Yu Q, Liu JH, et al. Src family protein-tyrosine kinases alter the function of PTEN to regulate phosphatidylinositol 3-kinase/AKT cascades. J Biol Chem 2003;278:40057-66. [PubMed]
- Chong CR, Jänne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med 2013;19:1389-400. [PubMed]
- Salt MB, Bandyopadhyay S, McCormick F. Epithelial-to-Mesenchymal Transition Rewires the Molecular Path to PI3K-Dependent Proliferation. Cancer Discov 2014;4:186-99. [PubMed]
- Niederst MJ, Benes CH. EMT Twists the Road to PI3K. Cancer Discov 2014;4:149-51. [PubMed]


