
A comparison of EGFR mutation status in tissue and plasma cell-free DNA detected by ADx-ARMS in advanced lung adenocarcinoma patients
Introduction
Lung cancer is one of the leading causes of cancer-related deaths worldwide. Recently, the clinical management of patients with advanced non-small cell lung cancer (NSCLC) has shifted from a histology-based to a molecularly-driven approach because of the identification of actionable genetic alterations and the subsequent development of highly effective targeted therapies. Genomic analysis has become routine in clinical practice to identify the molecular predictors of targeted therapies efficacy, such as somatic epidermal growth factor receptor (EGFR) mutations (1-3). The EGFR-tyrosine kinase inhibitors (EGFR-TKIs) have been successfully developed, and demonstrated a much higher objective response rate (ORR), a prolonged progression-free survival (PFS), better quality of life (QoL), and to have fewer side effects compared to first-line of chemotherapy for the advanced NSCLC patients with sensitizing EGFR mutations in large randomized phase III clinical trials (4-6). Although tumor tissue is still recognized to be the preferred standard sample for EGFR mutation detection, it is not readily available from every patient and may suffer from spatial and temporal heterogeneity (7-10). Therefore, extensive research has explored EGFR mutation detection in the plasma cell-free plasma DNA (cfDNA), with the primary aims of developing a novel, non-invasive, dynamic diagnostic method, with the potential advantage of providing a longitudinal evaluation of patients response and resistance to treatment (11). However, the concordance rate of the EGFR mutations status between plasma and tumor tissue varies (12,13).
Previous studies have shown that there are different methods used to detect the EGFR mutation status in cfDNA including direct sequencing, amplification-refractory mutation system (ARMS), next-generation sequencing (NGS), droplet digital PCR (ddPCR) and others (9,14-16).
The ARMS has been widely used in the clinical setting to guide the use of targeted therapy, with the most significant feature of being simple to operate and having a short turn-around time (8,17). However, the accuracy of its diagnostic ability remains inconsistent. For example, one study demonstrated a sensitivity of 60% and a specificity of 97% (18), while another study reported an inconsistent value of only 48.2% and 95.4%, respectively (19).
The purpose of this study is to evaluate the performance of ADx-Amplification Refractory Mutation System (ADx-ARMS) for the detection of EGFR mutations in cfDNA plasma as well as the consistency between the EGFR mutations of the tumor tissues and plasma. Moreover, we also evaluated the clinical treatment outcomes according to the EGFR mutation status in advanced lung adenocarcinoma patients.
Methods
Study population and inclusion criteria
This prospective study was performed in the Department of Pulmonary and Critical Care Medicine, the First Affiliated Hospital of Wenzhou Medical University, a tertiary teaching hospital in Zhejiang, China. Patients with histologically confirmed advanced (stage IIIB/IV) lung adenocarcinoma according to the seventh edition of the TNM classification of the malignant tumors system (20) from 01 April 2016 to 01 January 2017 were enrolled in this study. Criteria included newly diagnosed or having a progressive disease (PD) after EGFR-TKIs or recurrence after surgery without subsequent treatment. Patients without valid tissue or plasma samples or with an interval between the collection of tissue and the paired plasma sample longer than 14 days were excluded.
This study was approved by the institutional review board of the First Affiliated Hospital of Wenzhou Medical University (No. 2016017). All patients enrolled in this study signed an informed consent form.
Specimen collection and DNA extraction
The tumor tissues and paired peripheral blood (10 mL) from each patient were collected before subsequent treatment. Four to eight sections (5 mm thickness) of formalin-fixed, paraffin-embedded (FFPE) diagnostic tumor tissue samples were used for the DNA extraction. Peripheral blood samples were collected into EDTA tubes and centrifuged at 2,000 g for 10 min and then again for 8,000 g at 20 °C for 10 min. Plasma was separated within 1 hour and stored at a temperature of −80 °C until the cfDNA extraction time. The tumor tissue from the DNA was isolated from the FFPE slides using a QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Plasma cfDNA was extracted from 2 mL of the plasma from each patient using the QIAamp Circulating Nucleic Acid kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. EGFR mutations were immediately detected using the eluted DNA.
EGFR-sensitive mutation detection by ADx-ARMS EGFR kit
Complying with the manufacturer’s instructions, the EGFR mutations of the tumor tissues and plasma cfDNA samples were detected by using an ADx-ARMS kit (Amoy Diagnostics, Xiamen, China) (21) which had been approved by the Chinese Food and Drug Administration in 2010, to detect the 29 most-common EGFR mutations described in lung cancer (Table S1). The data analysis followed the manufacturer’s instructions as well.
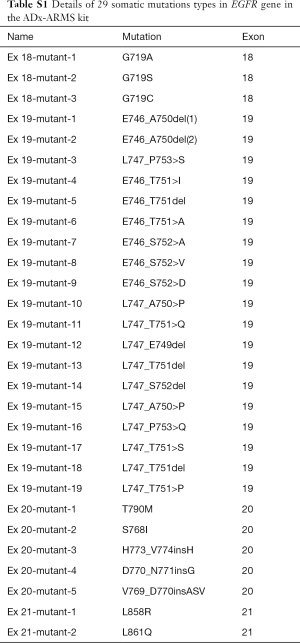
Full table
The NGS analysis for genomic DNA from plasma was performed in patients with an inconsistent gene region mutation in the paired plasma and tissues by the ADx-ARMS. DNA was processed using an AmoyDx Panel of 10 genes (Amoy Diagnostics, Xiamen, China) that are of clinical interest for lung cancer. Genomic DNA NGS library was captured for target enrichment and sequenced on the Illumina Nextseq 500 system (Illumina, San Diego, CA, USA) using the Nextseq 500 High Output kit v2 (300 cycles). Experimental procedure and data analysis followed the manufacturer’s instructions in detail.
Statistical analysis
Data were analyzed by SPSS Statistics version 23.0. EGFR mutations in the tissue samples were recognized as the standard when calculating the sensitivity and specificity for comparing plasma, while the consistency was measured using a Chi-squared test. We assessed the predictive value of the plasma cfDNA EGFR mutations for treatment outcomes with the ORR and PFS according to the Response Evaluation Criteria in Solid Tumors version 1.1 (22) and using the Kaplan-Meier method and the log-rank test, respectively. The data cutoff date used for survival analysis is 31 January 2018. All P values are two-sided and confidence intervals are at the 95% level, with statistical significance defined as P≤0.05.
Results
Patient characteristics
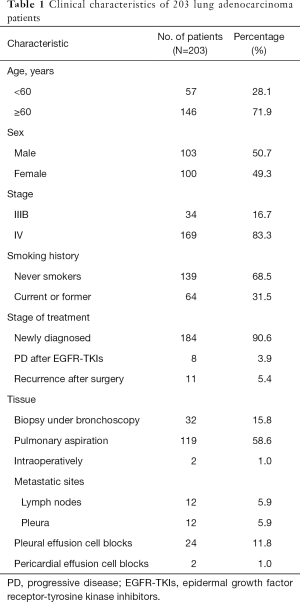
In total, 268 patients were enrolled in this study. Thirty-nine patients without a paired tumor tissue and 26 patients with more than 14 days interval between the collection of tissue and matched blood specimens were excluded from the analysis. Finally, 203 patients entered the analysis, which consisted of 100 women and 103 men. Sixty-four (31.5%) patients were smokers, 184 (90.6%) patients were newly diagnosed with advanced NSCLC, and 169 (83.3%) patients were classified as stage IV. From these FFPE tissue samples, 119 (58.6%) were collected via computed tomography-guided percutaneous lung biopsies, and 24 (11.8%) were pleural effusion cell blocks. All specimens were confirmed by histopathology with the diagnosis of lung adenocarcinomas. The clinical characteristics of these patients are shown in Table 1.
Full table
EGFR mutations in tumor tissue and plasma samples
EGFR mutations were detected in 119 (58.6%, 119/203) of the 203 tumor tissue samples, of which 58 (48.7%, 58/119) samples detected a single EGFR exon 19 deletion (19-Del), 46 (38.7%, 46/119) samples single EGFR L858R mutations, 2 (1.7%, 2/119) samples EGFR G719X mutations, 2 (1.7%, 2/119) samples EGFR 20-ins mutations, 2 (1.7%, 2/119) samples EGFR L861Q mutations, and 1 (0.8%, 1/119) sample EGFR S768I mutations. Eight (6.7%, 8/119) tumor tissue samples had detected multiple mutations (4 carried both 19 E19Del + T790M mutations; 4 carried both L858R + T790M mutations).
EGFR mutations were detected in 64 (31.5%, 64/203) plasma samples using the ADx-ARMS, including 33 (51.5%, 33/64) EGFR 19-Del, 27 (42.2%, 27/64) EGFR L858R mutations, 2 (3.1%, 2/64) EGFR 20-ins mutations, 1 (1.6%, 1/64) EGFR L861Q mutation, and 1 (1.6%, 1/64) EGFR S768I mutation. The details of the EGFR mutation status in tumor tissues and the plasma samples are shown in Table S2.
Full table
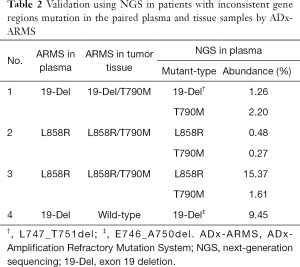
Fifty-six patients with documented EGFR mutation in tumor tissue had no detectable EGFR mutation in the paired plasma cfDNA test using ADx-ARMS. One patient with plasma EGFR mutation had no detectable mutation in the corresponding tumor tissue sample. Three cases carried the EGFR mutation in both tumor tissues, and matched plasma samples, but were inconsistent in the specific gene regions. One carried both 19-Del + T790M in tumor tissue but carried single 19-Del in plasma, and 2 carried both L858R + T790M but single L858R in plasma. We validated these 4 cases using NGS in plasma. Two of them carried L858R + T790M and one carried 19-Del + T790M, consistent with the result of tumor tissue detected by ADx-ARMS. The other one carried 19-Del, consistent with the result of ADx-ARMS in plasma. The details of the EGFR mutation status in tumor tissues and the plasma samples using NGS are listed in Table 2.
Full table
The sensitivity, specificity and consistency of detecting EGFR mutation by ARMS
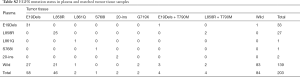
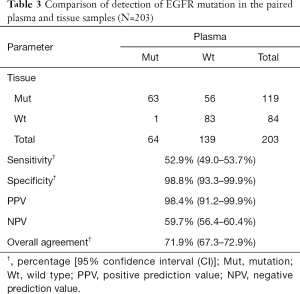
As compared to the paired tumor tissues, the sensitivity and specificity for EGFR mutation detection in plasma by ADx-ARMS was at 52.9% [with a 95% confidence interval (CI), 49.0–53.7%) and 98.8% (95% CI, 93.3–99.9%), respectively]. The PPV was 98.4% (95% CI, 91.2–99.9%) and the NPV was 59.7% (95% CI, 56.4–60.4%). The EGFR mutations in the tissue and the paired plasma are summarized in Table 3. Of the 203 matched samples, 146 presented consistent EGFR mutation status in the plasma and the paired tumor tissues using the ARMS. The overall consistency of the EGFR mutation was at 71.9%. Moreover, the kappa test showed a coefficient value of 0.472 revealing the degree of agreement for detecting EGFR mutations in tissues and plasma.
Full table
EGFR mutations in tumor tissue or plasma for predicting efficacy of EGFR-TKIs treatment
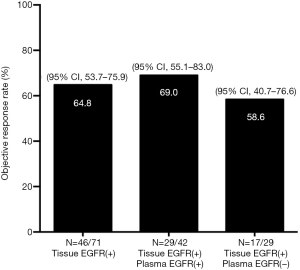
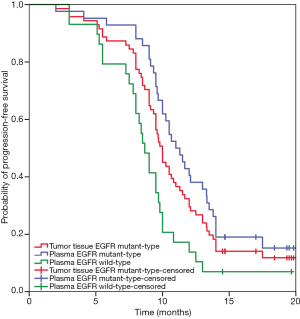
Seventy-one patients of the total 119 patients with the EGFR mutation in their tumor tissues had received a Gefitinib treatment [Iressa(R), Astra Zeneca Inc., UK] or an Icotinib treatment (Zhejiang Bata Pharma Ltd., China) as their first-line of treatment. Forty-six (64.8%) patients achieved a partial response (PR), 23 (32.4%) had a sustained disease response (SD), and 2 (2.8%) had PD. At data cut-off, 62 (87.3%) patients experienced PD and none of the patients had died. The ORR was 64.8% (95% CI, 53.7–75.9%). An objective response was seen in 42 patients who were EGFR mutation-positive in their plasma as detected by the ADx-ARMS, with an ORR of 69.0% (95% CI, 55.1–83.0%) (29 with PR, 13 with SD). The ORR was 58.6% (95% CI, 40.7–76.6%) for 29 patients with EGFR wild-type in their plasma. There were no significant differences between the three subgroups (P=0.664) (Figure 1). The median PFS of the patients with EGFR mutations detected by the ADx-ARMS in their tumor tissue and plasma samples was 10.0 months (95% CI, 9.3–10.7 months) vs. 11.0 months (95% CI, 9.7–12.3 months) (P=0.175), while the median PFS was 8.7 months (95% CI, 7.8–9.5 months) for the patients with an EGFR wild-type in their plasma, which was significantly shorter when compared to patients with an EGFR mutant type (P=0.001). The Kaplan-Meier curves for the PFS according to the tissue and plasma mutation status are shown in Figure 2.
Discussion
Plasma cfDNA has been proposed as an alternative surrogate for the detection of the EGFR mutation because it is feasible and sensitive (23). In the past decades, many methods have been developed to detect EGFR mutations using cfDNA level in plasma samples (16,24-27). A meta-analysis indicated that the pooled sensitivity for cfDNA was 67.4% and the specificity was 93.5% (28). Compared with a long turn-around time, a complicated procedure, and a limitation of the equipment availability of NGS (29), the ARMS assay has been found to be a stable and easy-to-operate method for assessing the EGFR mutation status for routine clinical use. Previous studies have reported that the ARMS achieved a 48.2–67.4% sensitivity and a 93.5–100% specificity when testing plasma when compared with the matched tumor tissues for the EGFR mutation status (8,18,28,30,31). In this study, we chose the ADx-ARMS method with the analytical sensitivity of 1% to evaluate the paired tumor tissue samples. We found a moderate sensitivity of 52.9%, but a high specificity of 98.8% for detecting EGFR mutations in plasma by ADx-ARMS, consistent with the previous studies (18,19) which have suggested that EGFR mutations detected in the plasma might be highly predictive of identical mutations in a paired tumor.
In the present study, only three patients had exhibited inconsistent EGFR mutations between their tumor tissue and the corresponding plasma samples. Validated by the NGS, the EGFR mutation status of these three patients was validated according to the detection results of the ADx-ARMS for tumor tissue. The relatively low abundance of the T790M mutation (0.27–2.2%) which has been observed by NGS might contribute to the false negative of the plasma T790M detection by the ADx-ARMS whose detection limit for the gene mutation was 1%. Apart from the sensitivity of the method itself, it has been reported that the low concentration of cfDNA extracted from plasma samples could lead to the low input of cfDNA template into the reaction, thus affecting EGFR mutation detection (32). Additionally, prolonged storage of blood samples could influence the detection efficiency of the EGFR mutation in the plasma as well (19). Generally, a high quantity of T790M predicted an inferior PFS after EGFR-TKI when it was compared with the low ones (33). In our study, two out of three carried de novo EGFR T790M mutations and the other one was an acquired resistance to the EGFR-TKIs therapy. All of them did not receive subsequent treatment after EGFR-TKIs and were alive at data cut-off, so we could not perform an overall survival outcome analysis.
In contrast, there was one patient who carried 19-Del in the plasma as detected by ADx-ARMS but was negative in the paired tumor tissue. This result was validated by NGS which was consistent with the result of ARMS testing in the plasma. The intra-tumor heterogeneity of the genetic abnormalities might be a possible reason for this inconsistency. The heterogeneity shows a potential advantage for combining the detection of the EGFR status in the plasma with the detection of EGFR status in tissue, addressing the issue of intra-tumor heterogeneity (34). Importantly, although the number of patients that tested positive for EGFR mutation in plasma but negative in tissues was limited, these subsets of patients might lose the chance of receiving anti-EGFR targeted therapy. Therefore, a combination of the detection of EGFR mutation status in both the tissue and the cfDNA might be clinically impactful (32).
Previous studies have indicated that T790M is the most common mechanism of acquired resistance to first- and second-generation EGFR-TKIs, accounting for approximately 60% of cases (35,36). In our cohort we found 8 patients who were T790M mutation positive in tumor tissues (4 with both 19-Del and T790M positive and 4 with both L858R and T790M positive). Among them, 6 carried de novo EGFR T790M mutation which was associated with primary resistance to treatment (37) and the other two were acquired resistance to the EGFR-TKIs therapy. However, none were detected for the T790M mutation by the ADx-ARMS in the plasma. As we know, plasma has become a preferred surrogate for tumor tissues, to explore the resistance mechanism and to implement a dynamic monitoring method when it is difficult to perform a re-biopsy. However, unlike the 19-Del or the L858R mutation, the T790M mutation is characterized by a low abundance and a high frequency, especially in the cfDNA (35). Yang et al. showed the sensitivity of detecting T790M in cfDNA by CastPCR was 45% (38). Our data might indicate the ADx-ARMS was not sensitive enough to detect the T790M mutation in the cfDNA as well (39-41). Since the majority of the patients enrolled in our study were newly diagnosed, additional clinical studies with PD after EGFR-TKIs and larger sample sizes are necessary to assess the performance of the ADx-ARMS for detecting a T790M mutation in the patients who had acquired resistance to the EGFR-TKIs.
Previous studies have reported significant correlations between the EGFR mutation status in plasma and the clinical response to EGFR-TKIs (33,42,43). According to the FASTACT-2 study, the patients with plasma cfDNA EGFR mutations had a significantly longer PFS when compared with patients without EGFR mutations in the plasma (13.1 vs. 6.2 months) (30). Another matched plasma and tumor samples study with DHPLC showed similar results of a prolonged PFS (11.1 vs. 5.9 months, P=0.044) and higher ORR rate of 62.2% with the EGFR mutations responding to Gefitinib treatment (43). Similarly, in the IFUM study, the ORRs of the baseline plasma EGFR mutation positive patients were better than in the negatives (76.9% vs. 59.5%) (8). In our study, the ORR with EGFR-TKIs in the group of patients with an EGFR mutation in plasma reported by the ADx-ARMS was 69.0%, higher than the ORR rate of 64.8% and 58.6% for the patients who were EGFR mutation-positive in tumor tissue and the patients who were EGFR mutation-negative in plasma, respectively. Patients with plasma cfDNA EGFR mutations also had longer median PFS than those with tumor tissue EGFR mutations and with no EGFR mutations in plasma (11.0 vs. 10.0 vs. 8.7 months). Patients with EGFR mutation in cfDNA tended to have a higher ORR and a longer median PFS, suggesting that the EGFR mutation in the plasma detected by the ADx-ARMS can predict the patients’ benefit from the EGFR-TKIs treatment, which is consistent with previously reported data (44,45). This is an interim analysis, therefore mature data for PFS and OS in the patients treated with EGFR-TKIs are not available yet. It is worth noting that most patients (98.6%, 70/71) included in the analysis of treatment outcomes are with EGFR sensitive mutations.
In conclusion, in this large prospective study we compared the EGFR mutation status in plasma cfDNA and the corresponding tumor tissues using ADx-ARMS in patients with advanced lung adenocarcinoma. EGFR mutation status assessment in plasma cfDNA by the ADx-ARMS was confirmed to be a non-invasive, readily accessible and high specific clinically available tool to optimize patients’ selection for treatment with EGFR-TKIs. Our data support the routine clinical application of this technology, which could be used as the initial complementary EGFR mutation detection along with tumor biopsy (32). ADx-ARMS is undoubtedly limited by a moderate sensitivity, however, higher sensitive technology such as ADx-SuperARMS are expected to further improve EGFR mutation status detection in patients with advanced NSCLC (21).
In summary, ADx-ARMS is an approach with a high specificity but a moderate sensitivity for detecting EGFR mutations in the plasma cfDNA of patients with advanced lung adenocarcinoma especially with newly-diagnosed NSCLC. EGFR mutation status in plasma cfDNA using ADx-ARMS can predict tumor response to EGFR-TKIs. The combination of testing EGFR mutation status in the tumor tissue and plasma should be performed to improve the outcome of our patients.
Acknowledgements
Thanks to all the patients who participated in this study.
Funding: This work was supported by Wenzhou Municipal Science and Technology Bureau (CN) (ZH2017001).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This study was approved by the institutional review board of the First Affiliated Hospital of Wenzhou Medical University (No. 2016017). All patients enrolled in this study signed an informed consent form.
References
- McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press 2015. Adv Nutr (Bethesda) 2016;7:418-9.
- Siegel RL, Miller K, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016;66:7-30. [Crossref] [PubMed]
- Perdigones N, Murtaza M. Capturing tumor heterogeneity and clonal evolution in solid cancers using circulating tumor DNA analysis. Pharmacol Ther 2017;174:22-6. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Goto K, Ichinose Y, Ohe Y, et al. Epidermal growth factor receptor mutation status in circulating free DNA in serum: from IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol 2012;7:115-21. [Crossref] [PubMed]
- Shi Y, Zhang L, Liu X, et al. Icotinib versus gefitinib in previously treated advanced non-small-cell lung cancer (ICOGEN): a randomised, double-blind phase 3 non-inferiority trial. Lancet Oncol 2013;14:953-61. [Crossref] [PubMed]
- Kawamura T, Kenmotsu H, Taira T, et al. Rebiopsy for patients with non-small-cell lung cancer after epidermal growth factor receptor-tyrosine kinase inhibitor failure. Cancer Sci 2016;107:1001-5. [Crossref] [PubMed]
- Douillard JY, Ostoros G, Cobo M, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol 2014;9:1345-53. [Crossref] [PubMed]
- Xu F, Wu J, Xue C, et al. Comparison of different methods for detecting epidermal growth factor receptor mutations in peripheral blood and tumor tissue of non-small cell lung cancer as a predictor of response to gefitinib. Onco Targets Ther 2012;5:439-47. [PubMed]
- Errico A. Lung cancer: Heterogeneity in space and time. Nat Rev Clin Oncol 2014;11:684. [PubMed]
- Helman E, Nguyen M, Karlovich CA, et al. Cell-Free DNA Next-Generation Sequencing Prediction of Response and Resistance to Third-Generation EGFR Inhibitor. Clin Lung Cancer 2018;19:518-30.e7. [Crossref] [PubMed]
- Esposito A, Bardelli A, Criscitiello C, et al. Monitoring tumor-derived cell-free DNA in patients with solid tumors: clinical perspectives and research opportunities. Cancer Treat Rev 2014;40:648-55. [Crossref] [PubMed]
- Wan JC, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. [Crossref] [PubMed]
- Aravanis AM, Lee M, Klausner RD. Next-Generation Sequencing of Circulating Tumor DNA for Early Cancer Detection. Cell 2017;168:571-4. [Crossref] [PubMed]
- Zhu G, Ye X, Dong Z, et al. Highly Sensitive Droplet Digital PCR Method for Detection of EGFR-Activating Mutations in Plasma Cell-Free DNA from Patients with Advanced Non-Small Cell Lung Cancer. J Mol Diagn 2015;17:265-72. [Crossref] [PubMed]
- Weber B, Meldgaard P, Hager H, et al. Detection of EGFR mutations in plasma and biopsies from non-small cell lung cancer patients by allele-specific PCR assays. BMC cancer 2014;14:294. [Crossref] [PubMed]
- Liu X, Lu Y, Zhu G, et al. The diagnostic accuracy of pleural effusion and plasma samples versus tumour tissue for detection of EGFR mutation in patients with advanced non-small cell lung cancer: comparison of methodologies. J Clin Pathol 2013;66:1065-9. [Crossref] [PubMed]
- Ma M, Shi C, Qian J, et al. Comparison of plasma and tissue samples in epidermal growth factor receptor mutation by ARMS in advanced non-small cell lung cancer. Gene 2016;591:58-64. [Crossref] [PubMed]
- Li X, Ren R, Ren S, et al. Peripheral blood for epidermal growth factor receptor mutation detection in non-small cell lung cancer patients. Transl Oncol 2014;7:341-8. [Crossref] [PubMed]
- Lababede O, Meziane M, Rice T. Seventh edition of the cancer staging manual and stage grouping of lung cancer: quick reference chart and diagrams. Chest 2011;139:183-9.
- Li Y, Xu H, Su S, et al. Clinical validation of a highly sensitive assay to detect EGFR mutations in plasma cell-free DNA from patients with advanced lung adenocarcinoma. PloS One 2017;12:e0183331. [Crossref] [PubMed]
- Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. [Crossref] [PubMed]
- Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov 2014;4:650-61. [Crossref] [PubMed]
- Uchida J, Kato K, Kukita Y, et al. Diagnostic Accuracy of Noninvasive Genotyping of EGFR in Lung Cancer Patients by Deep Sequencing of Plasma Cell-Free DNA. Clin Chem 2015;61:1191-6. [Crossref] [PubMed]
- Taniguchi K, Uchida J, Nishino K, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res 2011;17:7808-15. [Crossref] [PubMed]
- Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 2014;20:1698-705. [Crossref] [PubMed]
- Kim HR, Lee SY, Hyun DS, et al. Detection of EGFR mutations in circulating free DNA by PNA-mediated PCR clamping. J Exp Clin Cancer Res 2013;32:50. [Crossref] [PubMed]
- Luo J, Shen L, Zheng D. Diagnostic value of circulating free DNA for the detection of EGFR mutation status in NSCLC: a systematic review and meta-analysis. Sci Rep 2014;4:6269. [Crossref] [PubMed]
- Haimovich AD. Methods, challenges, and promise of next-generation sequencing in cancer biology. Yale J Biol Med 2011;84:439-46. [PubMed]
- Mok T, Wu Y, Lee J, et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res 2015;21:3196-203. [Crossref] [PubMed]
- Qian X, Liu J, Sun Y, et al. Circulating cell-free DNA has a high degree of specificity to detect exon 19 deletions and the single-point substitution mutation L858R in non-small cell lung cancer. Oncotarget 2016;7:29154-65. [Crossref] [PubMed]
- Wan R, Wang Z, Lee JJ, et al. Comprehensive Analysis of the Discordance of EGFR Mutation Status between Tumor Tissues and Matched Circulating Tumor DNA in Advanced Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:1376-87. [Crossref] [PubMed]
- Wang Z, Chen R, Wang S, et al. Quantification and dynamic monitoring of EGFR T790M in plasma cell-free DNA by digital PCR for prognosis of EGFR-TKI treatment in advanced NSCLC. PloS One 2014;9:e110780. [Crossref] [PubMed]
- Dong N, Shi L, Wang DC, et al. Role of epigenetics in lung cancer heterogeneity and clinical implication. Semin Cell Dev Biol 2017;64:18-25. [Crossref] [PubMed]
- Zhu L, Zhang S, Xun Y, et al. Comparison of the Amplification Refractory Mutation System, Super Amplification Refractory Mutation System, and Droplet Digital PCR for T790 M Mutation Detection in Non-small Cell Lung Cancer after Failure of Tyrosine Kinase Inhibitor Treatment. Pathol Oncol Res 2018;24:843-51. [Crossref] [PubMed]
- Cheng X, Chen H. Tumor heterogeneity and resistance to EGFR-targeted therapy in advanced nonsmall cell lung cancer: challenges and perspectives. Onco Targets Ther 2014;7:1689-704. [Crossref] [PubMed]
- Su KY, Chen HY, Li KC, et al. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J Clin Oncol 2012;30:433-40. [Crossref] [PubMed]
- Yang Y, Shen X, Li R, et al. The detection and significance of EGFR and BRAF in cell-free DNA of peripheral blood in NSCLC. Oncotarget 2017;8:49773-82. [PubMed]
- Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol 2016;34:3375-82. [Crossref] [PubMed]
- Lee Y, Lee GK, Lee YS, et al. Clinical outcome according to the level of preexisting epidermal growth factor receptor T790M mutation in patients with lung cancer harboring sensitive epidermal growth factor receptor mutations. Cancer 2014;120:2090-8. [Crossref] [PubMed]
- Sorensen BS, Wu L, Wei W, et al. Monitoring of epidermal growth factor receptor tyrosine kinase inhibitor-sensitizing and resistance mutations in the plasma DNA of patients with advanced non-small cell lung cancer during treatment with erlotinib. Cancer 2014;120:3896-901. [Crossref] [PubMed]
- Mitsudomi T, Kosaka T, Endoh H, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol 2005;23:2513-20. [Crossref] [PubMed]
- Bai H, Mao L, Wang H, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol 2009;27:2653-9. [Crossref] [PubMed]
- Kimura H, Suminoe M, Kasahara K, et al. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA). Br J Cancer 2007;97:778-84. [Crossref] [PubMed]
- Kimura H, Kasahara K, Kawaishi M, et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non-small-cell lung cancer. Clin Cancer Res 2006;12:3915-21. [Crossref] [PubMed]


