
The role of the gut microbiome on the efficacy of immune checkpoint inhibitors in Japanese responder patients with advanced non-small cell lung cancer
Introduction
Cancer immunotherapy, such as programmed cell death protein 1 (PD-1)/programmed death-ligand 1 (PD-L1) checkpoint inhibitors, are currently being developed as a promising alternative strategy for the treatment of advanced non-small cell lung cancer (NSCLC) (1-4). Several mechanisms favoring the response to immune checkpoints have been reported, such as a high expression of PD-L1 (5). However, potent biomarkers that select candidates to have a favorable clinical response to immune checkpoint inhibitors (ICIs) have not been developed.
The gut microbiota, which forms a complex interaction with the human host through immunity and metabolism to maintain homeostasis, consists of 1,000 bacterial species and more than 100 trillion enteric bacteria (6,7). Recent clinical trials have shown the relationship between the effects of an anti-PD-1 antibody and the composition of the gut microbiome in malignant melanoma and NSCLC (8-11). Therefore, the gut microbiome has pivotal roles in the prediction of response to ICI treatment in NSCLC patients. In this study, we retrospectively analyzed the relationship between the efficacy of ICI treatment and the gut microbiome in Japanese patients with NSCLC who were administered ICI.
Methods
Patients
This was a retrospective study evaluating the effects of gut microbiota on the efficacy of ICIs in long-term responders with advanced NSCLC. We consecutively selected 17 Japanese patients with advanced NSCLC who were treated with ICIs for >3 months at the University Hospital Kyoto Prefectural University of Medicine in Japan between December 2015 and March 2018, regardless of any previous treatment with cytotoxic chemotherapy. We collected stool samples one time per patient during ICI treatment between June 2017 and March 2018. We defined responder (R) (partial response to ICI treatment) or non-responder (NR) (stable or progressive disease after ICI treatment at the time of first clinical evaluation) according to the RECIST 1.1 evaluation. Radiological evaluation occurred at least every 12 weeks. The study protocol was approved by the Ethics Committees of Kyoto Prefectural University of Medicine (No. ERB-C-1126), and conducted in accordance with the principles of the Declaration of Helsinki. The TNM stage was classified using version 7 of the TNM stage classification system. Time to treatment failure (TTF) was defined as the time from the start of treatment to treatment discontinuation for any reason. Overall survival (OS) was defined as the treatment start time to death from any cause.
Tumor PD-L1 analysis
PD-L1 expression was analyzed at SRL, Inc., with the PD-L1 IHC 22C3 pharmDx assay (Agilent Technologies, Santa Clara, CA). The PD-L1 TPS (Tumor Proportion Score) was calculated as the percentage of at least 100 viable tumor cells for complete or partial membrane staining. The pathologists at the commercial vendor interpreted the TPS results.
Samples, DNA extraction, 16S rRNA gene amplification, sequencing, and data analysis
The gut microbiome analysis was carried out by MykinsoPro gut microbiome testing (Cykinso, Inc., Tokyo, Japan). DNA extraction from fecal samples was performed by an automated DNA extraction machine (GENE PREP STAR PI-480, Kurabo Industries Ltd, Osaka, Japan) according to the manufacturer's protocol. The V1-V2 region of the 16S rRNA gene was amplified by RT-PCR. To sequence the 16S amplicons by Illumina MiSeq platform, dual index adapters were attached using the Nextera XT Index kit. The sequence library preparations were performed according to the 16S library preparation protocol by Illumina (San Diego, CA). The libraries were sequenced using the MiSeq Reagent Kit v2 (500 Cycles, 250 bp paired-end). After clustering with 97% homology using UCLUST version 1.2.22q (Edgar, 2010), operational taxonomic units (OTUs) were generated. The representative sequences of the OTUs were collated against the full-length 16S gene database Greengenes (v13.8) to determine the identity and composition of the bacterial genera. The QIIME (v1.8.0) pipeline was used for analysis from the clustering step through alpha diversity. The linear discriminant analysis, combined with effect size measurements (LEfSe), were used to quantitate differential taxonomic and functional pathway abundance between the groups (R vs. NR).
Statistical analysis
Statistical analyses were performed using the EZR version 1.30 statistical software (12). All statistical tests were two-sided, and P<0.05 was regarded as statistically significant. The demographic characteristics were expressed as frequencies and percentages for the categorical variables and as medians and ranges for the continuous variables. The categorical variables were compared using Fisher’s exact test. The TTF and OS were calculated using the Kaplan-Meier method, and the differences were compared using the log-rank test. The alpha diversity metrics and relative abundance of the gut microbiomes were compared by Mann-Whitney tests.
Results
Patient characteristics
Six NSCLC patients responded to ICI treatment, while 11 patients did not respond (Figure 1). A total of 13 (76.5%) patients were male, and the median age of all patients was 70 years (range, 56–83 years). The performance statuses were 0 in 5 (29.4%), and 1 in 12 (70.6%) patients. No patient experienced a CR (0%), 6 patients had a PR (35.3%), 10 had an SD (58.8%), and 1 had a PD (5.9%) when treated with ICIs according to the RECIST criteria indicating a response rate of 35.3%. The median TTF was 270 days for all NSCLC patients. OS could not be assessed because most patients were alive at the end of the study. The median follow-up time was 622 days for all NSCLC patients. The baseline clinicopathological characteristics of the patients are summarized in Table 1.

Full table
Association between the gut microbiome and outcome of ICIs in NSCLC patients
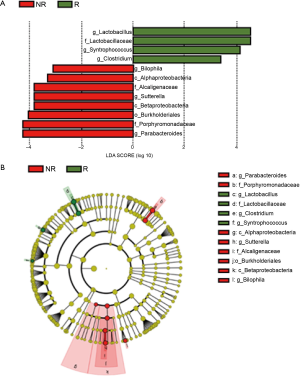
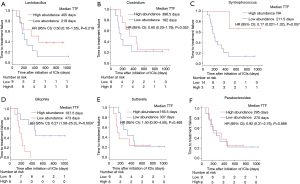
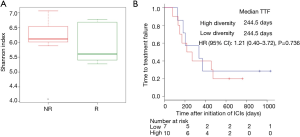
We performed 16S rRNA sequencing on the stool samples collected during ICI treatment and evaluated high dimensional class comparisons via LEfSe on the population of gut microbiota between the responders and non-responders of ICIs. The gut microbiomes in the ICI responders were significantly enriched with Lactobacillus, Clostridium, and Syntrophococcus when compared to those of non-responders. In contrast, the gut microbiomes of the ICI non-responders were significantly abundant with Bilophila, Sutterella, and Parabacteroides when compared to those of responders (Figure 2). Patients with a high abundance of Lactobacillus, Clostridium, and Syntrophococcus tended to have a longer TTF when compared to those with a lower abundance. Patients with a low amount of Bilophila and Sutterella had a significantly prolonged TTF than those with a high amount. However, there was no remarkable association between the abundance of Parabacteroides and TTF with ICIs (Figure 3). The alpha-diversity of the gut microbiota was not significantly different between the responders and non-responders and did not influence the TTF (Figure S1).
Discussion
In this study, we identified that the colonization of the operational taxonomic unit, including Lactobacillus and Clostridium, was associated with favorable clinical outcomes with ICI treatment. Lactobacillus has been reported to promote DC maturation and regulate host immunity in preclinical models (13). These observations suggest that the specific subpopulation enrichment of the gut microbiomes, such as Lactobacillus and Clostridium, may elicit T cell mobilization to the tumors, thereby enhancing the therapeutic effects of ICI treatment. In contrast, a subpopulation of the microbiome, such as Sutterella and Bilophila, was negatively associated with TTF with ICI treatment. These bacteria elicit inflammation within the tumor microenvironment, which may be related to the insensitivity to ICI treatment (14,15). Interestingly, Bilophila-induced inflammation and intestinal barrier dysfunctions have been shown to be suppressed by the probiotic, Lactobacillus, in mice models (14). Therefore, the management with probiotics may improve the response to maintenance therapy with ICIs. Further investigations are warranted to improve the clinical outcomes of NSCLC patients with ICIs.
This study has several limitations. First, it comprised a small retrospective sample and selected only patients who were undergoing ICI treatment for a long duration. Second, we selected patients who were being treated with ICIs for more than three months, which may cause a selection bias. Third, there may be a bias considering that we obtained data from our domestic hospital only. Forth, we evaluated the outcomes only once based on stool sampling during ICI treatment, and we have not evaluated the influence of ICI treatment on the gut microbiome. Therefore, a further prospective study is warranted to identify the role of the microbiome in evaluating the response to ICIs in NSCLC patients.
Conclusions
In summary, our observations suggest that the composition of the gut microbiome may be associated with therapeutic outcomes in patients with advanced NSCLC treated with ICIs for a long time. A further large-scale study is warranted to validate the composition of the gut microbiome as a novel clinical factor for the response to ICIs in NSCLC patients after a long duration.
Acknowledgments
Funding: This work was supported by the Kyoto Health Science Research Center (Kyoto, Japan).
Footnote
Conflicts of Interest: Dr. Yamada reports receiving research grants from Pfizer Inc., Ono Pharmaceutical Co., Ltd., Takeda Pharmaceutical Co. Ltd., and Chugai Pharmaceutical Co., Ltd. Dr. Uchino reports receiving research grants from Eli Lilly Japan K.K., AstraZeneca K.K., and Boehringer Ingelheim Japan Inc. Dr. Takayama reports receiving research grants from Chugai-Roche Co., and Ono Pharmaceutical Co., and personal fees from AstraZeneca Co., Chugai-Roche Co., MSD-Merck Co., Eli Lilly Co., Boehringer-Ingelheim Co., and Daiichi-Sankyo Co. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study protocol was approved by the Ethics Committees of Kyoto Prefectural University of Medicine and each hospital, and conducted in accordance with the principles of the Declaration of Helsinki. This was a retrospective study, so informed consent was waived.
References
- Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:1627-39. [Crossref] [PubMed]
- Brahmer J, Reckamp KL, Baas P, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 2015;373:123-35. [Crossref] [PubMed]
- Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540-50. [Crossref] [PubMed]
- Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017;389:255-65. [Crossref] [PubMed]
- Sacher AG, Gandhi L. Biomarkers for the Clinical Use of PD-1/PD-L1 Inhibitors in Non-Small-Cell Lung Cancer: A Review. JAMA Oncol 2016;2:1217-22. [Crossref] [PubMed]
- Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell 2014;157:121-41. [Crossref] [PubMed]
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006;124:837-48. [Crossref] [PubMed]
- Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018;359:97-103. [Crossref] [PubMed]
- Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018;359:104-8. [Crossref] [PubMed]
- Jin Y, Dong H, Xia L, et al. The Diversity of Gut Microbiome is Associated With Favorable Responses to Anti-Programmed Death 1 Immunotherapy in Chinese Patients With NSCLC. J Thorac Oncol 2019;14:1378-89. [Crossref] [PubMed]
- Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018;359:91-7. [Crossref] [PubMed]
- Kanda Y. Investigation of the freely available easy-to-use software 'EZR' for medical statistics. Bone Marrow Transplant 2013;48:452-8. [Crossref] [PubMed]
- Elawadli I, Brisbin JT, Mallard BA, et al. Differential effects of lactobacilli on activation and maturation of mouse dendritic cells. Benef Microbes 2014;5:323-34. [Crossref] [PubMed]
- Natividad JM, Lamas B, Pham HP, et al. Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat Commun 2018;9:2802. [Crossref] [PubMed]
- Hiippala K, Kainulainen V, Kalliomaki M, et al. Mucosal Prevalence and Interactions with the Epithelium Indicate Commensalism of Sutterella spp. Front Microbiol 2016;7:1706. [Crossref] [PubMed]


