Chromosomal rearrangements and their neoantigenic potential in mesothelioma
Introduction
Malignant pleural mesothelioma (MPM) is most commonly associated with exposure to the carcinogen asbestos, although some cases develop after therapeutic radiation or are inherited due to loss of function mutations in BRCA1 associated protein 1 (BAP1) (1,2). Consistent with the carcinogenicity of asbestos, previous studies frequently identified recurrent, structural chromosomal abnormalities in MPM using karyotyping (3). In contrast, more recent studies that used next-generation sequencing (NGS) to evaluate MPM reported a very low mutation burden overall (4,5). This finding was unexpected given that other carcinogen exposure associated tumors such as malignant melanoma and non-small cell lung cancer (NSCLC) are known to have a high mutation loads related to ultraviolet radiation and tobacco exposure respectively (6). A high tumor mutation burden is thought to be a surrogate for an increase in expression of tumor neoantigens that can be recognized by the immune system. In this context it has been demonstrated that patients with NSCLC with a high tumor mutation burden have superior progression-free survival compared to those with a low tumor mutation burden when treated with an immune checkpoint inhibitor that disrupts signaling between programmed cell death 1 ligand 1 (PD-L1) and its receptor PD-1 (7). Interestingly, despite reportedly very low tumor mutation burdens in patients with MPM, response rates and survival of patients with MPM treated with PD-1 inhibitors are similar to those of patients with NSCLC for second or later lines of therapy (8,9). Therefore, in our prior work we investigated the presence of structural chromosomal variations, not detected by conventional NGS techniques and their contribution to neoantigen expression in MPM (10).
Sequencing structural variants
While NGS is a powerful technique, based on its most common applications it is optimized for the detection of nucleotide variants. There are areas of the genome that are difficult to sequence (e.g., repetitive sequences), and genomic abnormalities that are not readily detected by standard NGS approaches (e.g., rearrangements) (11). Mate-pair sequencing (MPseq) differs from standard sequencing approaches namely in the library preparation step and bioinformatics analysis. For MPseq, large DNA fragments (>2 kb) are generated and their ends are labeled. These ends are then circularized, thus mating the ends of these longer fragments and giving the name mate-pair. These circularized fragments are then sheared, the mated ends are purified and sequenced as usual. Even though only a few hundred base pairs of sequence are generated, these results can identify structural events. Concordant reads map to a reference genome in the proper orientation and appropriate distance from one another, whereas discordant reads map closer or further to one another than expected and possibly with an incorrect orientation. Thus, chromosomal rearrangements (balanced and unbalanced), insertions and deletions can be robustly identified with MPseq.
Genomic array platforms may also be used to infer the presence of structural abnormalities; however, their ability to detect specific patterns can be limited. Whereas oscillating copy-number states might be identified, events involving multiple chromosomes will not likely be detected. Furthermore, prior work has shown that array platforms that consist of more than 250k probes are preferred for the detection of chromothripsis like events (12).
Structural abnormalities in mesothelioma
Recurrent chromosomal alterations such as losses of 1p and 9q were reported in MPM in prior karyotypic analyses (3). More recently, in order to resolve discrepancies in the detection of BAP1 mutations, a team of investigators identified that 3p21 (where BAP1 is located) is frequently involved with chromothripsis using a high-density comparative genomic hybridization array (13). These results suggested that standard NGS which detects nucleotide abnormalities cannot always detect structural abnormalities. Chromothripsis results from numerous double strand breaks of a large segment of a chromosome or a whole chromosome (Figure 1). The subsequent repair of these chromosomal breaks results in multiple novel rearrangements that are out of sequence from normal. The micronuclei that form around lagging chromosomes during cell division are not able to incorporate nuclear pore complexes and other non-core nuclear envelope proteins that protect chromosomes from fragmentation (14), and the loss of nuclear envelope integrity fosters chromosome fragmentation (15). Chromoplexy on the other hand represents a series of linked translocations, or a weaving of chromosomal fragments (Figure 1). Chromoplexy was first described in prostate cancer (16) and commonly involves the well-known TMPRSS2-ERG translocation (17). Chromoplexy likely results from the inappropriate ligation of chromosomal breaks of co-expressed loci; however, the inciting event is not well understood. Whereas chromothripsis typically involves a single chromosome, chromoplexy usually involves multiple chromosomes. These findings and statistical modeling support that chromothripsis and chromoplexy likely represent single, lineage-defining, catastrophic events during the evolution of a tumor (16). We previously reported that an ALK rearrangement in an inflammatory myofibroblastic tumor resulted from a complex chromoplectic rearrangement. Whereas that event involved the ALK oncogene (18), most of the rearrangements we have observed in MPM involve tumor suppressor genes and non-coding DNA (10). We currently do not know if the ALK rearrangements reported in peritoneal mesothelioma result from chromoplexy or other complex, structural events (19).
In order to resolve the reported discrepancy between the numerous chromosomal abnormalities detected by karyotyping and the low tumor mutation burden detected by standard exome sequencing in MPM we leveraged MPseq and RNAseq to detect transcribed structural variants. In a series of 22 MPM specimens, we found 1,535 chromosomal rearrangements that resulted in junctions or novel fusions of non-coding DNA or genes (median 41, range 3–298) (Figure 2). For comparison, prior work only identified 43 rearrangements in 211 specimens. We further characterized the abnormalities detected by MPseq and observed that many rearrangements were part of a pattern of chromoanagenesis consistent with chromothripsis (present in 10/22 specimens, 45%), chromoplexy (present in 9/22 specimens, 41%), or both (present in 7/22 specimens, 32%). In nine out of ten cases with chromothripsis, there were multiple chromothriptic events.
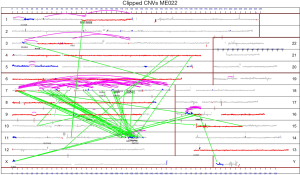
Although MPseq can robustly detect chromothripsis and chromoplexy, other techniques can infer the presence of chromothripsis. More specifically, chromothripsis was first identified in the malignant cells of a patient with chronic myeloid leukemia (20) and subsequent work screened for chromothripsis using single-nucleotide polymorphism array data (12,21). Accordingly, criteria for the inference of chromothripsis have been developed to aid in the appropriate identification of this event (Table 1) (22).
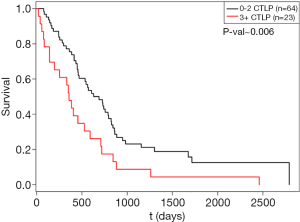
Since standard NGS approaches are not optimized to detect chromosomal rearrangements, the frequencies of chromothripsis and chromoplexy are likely underestimated in human cancers. Since chromothripsis results in oscillating copy number changes in a relatively small chromosomal region of high-quality genomic array data, methods have been developed to infer the presence of these events. CTLPScanner is a scan-statistic based algorithm that identifies clusters of copy number status changes called chromothripsis-like patterns (CTLPs) (23). We analyzed the copy number segmentation data of the mesothelioma cohort (n=87) in The Cancer Genome Atlas (TCGA) for chromothripsis using CTLPScanner (5). We used this approach since the DNA library preparation steps in TCGA were not compatible with MPseq. We identified that 69% of the mesothelioma specimens in TCGA had copy number changes consistent with chromothripsis. Subjects with 3 or more CTLPs had a significantly worse prognosis than those with 2 or fewer CTLPs (HR =2.002, P=0.006) (Figure 3). This analysis of CTLPs in the mesothelioma cohort of TCGA complements the results of our Mayo Clinic cohort with MPseq: both suggest that chromosomal rearrangements, and more specifically chromothripsis, are common in mesothelioma.
Neoantigenic potential of chromosomal rearrangements in mesothelioma
To determine whether transcription of chromosomal rearrangement-related junctions have neoantigenic potential, we used NetMHC-4.0 to predict whether any of the expressed junctions could encode peptides that could be presented by a patient’s HLA molecules (24,25). NetMHC uses artificial neural networks to predict the binding of peptides to MHC class I molecules. In one case of MPM (ME018) a total of 1,146 9mer, 10mer, and 11mer peptides were identified from 6 protein products of expressed chromosomal rearrangements with at least 10 RNAseq reads. Of these peptides, 75 were predicted to bind to that patient’s specific HLA molecules (HLA-A*02:05, HLA-A*24:02, HLA-B*15:01 and HLA-B*35:01), yielding 87 distinct epitopes. To evaluate whether the candidate rearrangement-related peptides with neoantigenic potential bind patient-specific HLA molecules, the 9mers of two candidate peptides (NYLETTSDF, CYGETYQNI) and the 11mer (NYLETTSDFHF) of one candidate peptide were selected based on their predicted immunogenicity. These candidate peptides resulted from the rearrangement of non-coding DNA with CADPS2 or non-coding DNA with NOD1. An MHC-peptide competitive binding assay was performed and demonstrated that all three peptides bound HLA-A*24:02 very well. These results suggest that the predicted peptide-MHS class I binding affinities were accurate.
To determine the effects of chromosomal rearrangements on tumor-infiltrating T cells, we used T cell receptor sequencing. We found a significant correlation between the number of T cell clones and predicted chromosomal rearrangement-related neoantigens. For each specimen we also calculated a productive clonality score which provides a measurement of clonal distribution. Pielou’s evenness index is used to derive the productive clonality score (26). In regards to T cell clones, Pielou’s evenness index scores the relative abundance (number of detected copies) of each clone. Productive clonality is the inverse of Pielou’s Evenness Index and ranges from 0 to 1, where 0 represents an even distribution of clones and 1 represents an uneven distribution of clones (aka clonal expansion of one or a few T cell clones). We detected a positive correlation between productive clonality with chromosomal rearrangements, rearrangements involving genes and predicted neoantigens. In other words, an increase in predicted neoantigens resulting from chromosomal rearrangements was correlated with clonal expansion of tumor-infiltrating T cells.
To determine whether there are patient T cells responsive to these predicted neoantigens we prospectively collected the peripheral blood mononuclear cells (PBMCs) and tumor from an additional patient who underwent surgery for mesothelioma at our institution. The tumor was analyzed as per the prior 22 cases and 86 potential neoantigens were predicted from the chromosomal rearrangements. We then synthesized eight peptides of these potential neoantigens that correspond to 20 epitopes (cross-presented on 1–4 patient HLA alleles, with a median of 2 neo-epitopes per peptide) for an interferon-γ enzyme-linked immunospot assay. This assay demonstrated that T cells responsive to these predicted neoantigens were present in this patient’s PBMCs. These results are the first to our knowledge that suggest chromosomal rearrangements have neoantigenic potential (Figure 4).
Beyond mesothelioma
The prevalence of chromothripsis is likely higher across malignancies than previously reported. With the use of low-density array platforms, it was suggested that chromothripsis was present in 1–5% of cancers (21). This rate was observed using CTLPs in a more recent study of 402 GEO series, encompassing 22,347 high quality genomic arrays of human cancer samples. CLTPs were identified in 918 cases and involved 1,269 chromosomes (12). These events were not related to chromosome size and were most commonly observed on chromosome 17. Also, TP53 mutations were more common in cases with CTLP events than without, although CTLP events were present in the absence of TP53 mutations. Furthermore, among the patients with available survival information, CTLP events were associated with worse outcomes than patients without CTLP events. Other approaches using whole genome sequencing with single nucleotide resolution have suggested that chromothripsis can be detected in 50% or more of several cancer types (27).
In regards to non-small cell lung cancer in which subsets of patients have chromosomal rearrangements that can be targeted therapeutically, a recent analysis of 138 cases of adenocarcinoma of the lung identified that 74% of the fusions of known oncogenes were related to chromothripsis or chromoplexy (28). These results suggest that complicated chromosomal rearrangements may play a significant role in oncogenesis. Whereas prior version of “Signatures of Mutational Processes in Human Cancer” did not include chromosomal rearrangements, the latest version recognizes these events (6).
Chromothripsis has also been identified in a patient with severe congenital abnormalities (29). Multiple de novo breakpoints on chromosomes 1, 4 and 10 were identified in this patient and were thought to be related to this patient’s congenital abnormalities. There are not many reports in the literature of chromoanagenesis in germline DNA; however, the application of sequencing techniques that can detect rearrangements may help identify congenital abnormalities when standard approaches fail.
Conclusions
Despite a low tumor mutation burden determined by single nucleotide changes, MPM is commonly characterized by unique chromosomal rearrangements that often fit a complex pattern of chromoanagenesis. Standard NGS sequencing strategies, optimized to detect nucleotide variants, are sub-optimal for the detection of these events while MPseq provides an excellent means for their detection. These chromosomal rearrangements can result in novel, unique gene junctions that can be expressed and potentially result in the presentation of several neoantigens. These predicted neoantigens can be presented by tumors on major histocompatibility complex (MHC) proteins and are correlated with clonal expansion of tumor infiltrating T cells. T cells responsive to these neoantigens have been identified in the circulation of a patient. These findings may have several implications for the use of immunotherapy. The predictive values of NGS-based tumor mutation burden measurements may be significantly enhanced by the addition of MPseq and the identification of rearrangement associated neo-antigens. Furthermore, rearrangement associated neo-antigens may also represent valuable targets for future anti-tumor vaccine strategies. Finally, chromosomal rearrangements are now recognized as a mutation signature in cancer and these events are likely to be important in the oncogenesis and immune recognition of not only in MPM but multiple malignancies including non-small cell lung cancer.
Acknowledgments
We would like to acknowledge BobbiAnn Jebens for her assistance with this manuscript.
Funding: None.
Footnote
Conflicts of Interest: Honoraria have been provided to Dr. Mansfield’s institution for participation on advisory boards for Abbvie, BMS and Genentech. Dr. Mansfield has received funding from BMS, Novartis, Verily and NIH. Dr. Mansfield is a non-remunerated member of the Board of Directors of the Mesothelioma Applied Research Foundation.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Carbone M, Adusumilli PS, Alexander HR Jr, et al. Mesothelioma: Scientific clues for prevention, diagnosis, and therapy. CA Cancer J Clin 2019;69:402-9. [Crossref] [PubMed]
- Testa JR, Cheung M, Pei J, et al. Germline BAP1 mutations predispose to malignant mesothelioma. Nat Genet 2011;43:1022-5. [Crossref] [PubMed]
- Popescu NC, Chahinian AP, DiPaolo JA. Nonrandom chromosome alterations in human malignant mesothelioma. Cancer Res 1988;48:142-7. [PubMed]
- Bueno R, Stawiski EW, Goldstein LD, et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat Genet 2016;48:407-16. [Crossref] [PubMed]
- Hmeljak J, Sanchez-Vega F, Hoadley KA, et al. Integrative Molecular Characterization of Malignant Pleural Mesothelioma. Cancer Discov 2018;8:1548-65. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Carbone DP, Reck M, Paz-Ares L, et al. First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2415-26. [Crossref] [PubMed]
- Alley EW, Lopez J, Santoro A, et al. Clinical safety and activity of pembrolizumab in patients with malignant pleural mesothelioma (KEYNOTE-028): preliminary results from a non-randomised, open-label, phase 1b trial. Lancet Oncol 2017;18:623-30. [Crossref] [PubMed]
- Mansfield AS. Immune checkpoint inhibition in malignant mesothelioma: does it have a future? Lung Cancer 2017;105:49-51. [Crossref] [PubMed]
- Mansfield AS, Peikert T, Smadbeck JB, et al. Neoantigenic Potential of Complex Chromosomal Rearrangements in Mesothelioma. J Thorac Oncol 2019;14:276-87. [Crossref] [PubMed]
- Adams DR, Eng CM. Next-Generation Sequencing to Diagnose Suspected Genetic Disorders. N Engl J Med 2018;379:1353-62. [Crossref] [PubMed]
- Cai H, Kumar N, Bagheri HC, et al. Chromothripsis-like patterns are recurring but heterogeneously distributed features in a survey of 22,347 cancer genome screens. BMC Genomics 2014;15:82. [Crossref] [PubMed]
- Yoshikawa Y, Emi M, Hashimoto-Tamaoki T, et al. High-density array-CGH with targeted NGS unmask multiple noncontiguous minute deletions on chromosome 3p21 in mesothelioma. Proc Natl Acad Sci U S A 2016;113:13432-7. [Crossref] [PubMed]
- Liu S, Kwon M, Mannino M, et al. Nuclear envelope assembly defects link mitotic errors to chromothripsis. Nature 2018;561:551-5. [Crossref] [PubMed]
- Hatch EM, Fischer AH, Deerinck TJ, et al. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 2013;154:47-60. [Crossref] [PubMed]
- Baca SC, Prandi D, Lawrence MS, et al. Punctuated evolution of prostate cancer genomes. Cell 2013;153:666-77. [Crossref] [PubMed]
- Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005;310:644-8. [Crossref] [PubMed]
- Mansfield AS, Murphy SJ, Harris FR, et al. Chromoplectic TPM3-ALK rearrangement in a patient with inflammatory myofibroblastic tumor who responded to ceritinib after progression on crizotinib. Ann Oncol 2016;27:2111-7. [Crossref] [PubMed]
- Hung YP, Dong F, Watkins JC, et al. Identification of ALK Rearrangements in Malignant Peritoneal Mesothelioma. JAMA Oncol 2018;4:235-8. [Crossref] [PubMed]
- Stephens PJ, Greenman CD, Fu B, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 2011;144:27-40. [Crossref] [PubMed]
- Kim TM, Xi R, Luquette LJ, et al. Functional genomic analysis of chromosomal aberrations in a compendium of 8000 cancer genomes. Genome Res 2013;23:217-27. [Crossref] [PubMed]
- Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell 2013;152:1226-36. [Crossref] [PubMed]
- Yang J, Liu J, Ouyang L, et al. CTLPScanner: a web server for chromothripsis-like pattern detection. Nucleic Acids Res 2016;44:W252-8. [Crossref] [PubMed]
- Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 2016;32:511-7. [Crossref] [PubMed]
- Nielsen M, Lundegaard C, Worning P, et al. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci 2003;12:1007-17. [Crossref] [PubMed]
- Pielou EC. Species-diversity and pattern-diversity in the study of ecological succession. J Theor Biol 1966;10:370-83. [Crossref] [PubMed]
- Cortés-Ciriano I, Lee JK, Xi R, et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. bioRxiv 2018.333617.
- Lee JJ, Park S, Park H, et al. Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma. Cell 2019;177:1842-57 e21.
- Kloosterman WP, Guryev V, van Roosmalen M, et al. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum Mol Genet 2011;20:1916-24. [Crossref] [PubMed]