
Presence of allele frequency heterogeneity defined by ctDNA profiling predicts unfavorable overall survival of NSCLC
Introduction
Recent studies have reported that the presence of subclonal mutations may compromise the anti-tumor immune response (1,2). These subclones may account for the genetic heterogeneity of tumor as well as resistance to treatment (3). As generation of subclones is driven by tumor mutations, the mutation distribution might serve as a surrogate measurement of subclones. Detection of mutations by circulating tumor DNA (ctDNA) has been increasingly accessible worldwide (4-7), and provides a comprehensive landscape across cancerous tissue for making it an ideal approach to assess the tumor heterogeneity. The variant allele fraction of different genetic mutations in ctDNA are often used to represent the clonal makeup of a tumor and high allele fraction mutations are more likely to be clonal, whereas lower values are more likely to be subclonal (8). Therefore, it is of great interest to explore the impact of distribution of mutation abundance on prognosis.
We hypothesized that the relatively low-frequency mutations in ctDNA might indicate allele frequency heterogeneity (AFH); a low ratio of allele frequency (AF) of a mutation to the maximum-somatic-allele-frequency (MSAF) was used to define the presence of AFH. The primary objective was, first, to explore whether AF/MSAF ratio that was below a defined cutoff point predicted prognosis of advanced NSCLC patients and, second, to evaluate the prognostic role of AFH (defined by the optimal AF/MSAF ratio) despite which drug the patients used.
Methods
Genomic and clinical data of advanced NSCLC patients from POPLAR (9) (N=211, NCT01903993) and OAK (10) (N=642, NCT02008227) trials from public database (https://doi.org/10.1038/s41591-018-0134-3) were included for primary analysis. Both POPLAR (randomized phase II trial) and OAK (randomized phase III trial) were designed to compared single agent atezolizumab with docetaxel as second/third-line therapy for patients with advanced NSCLC, and genomic alterations of pre-treatment plasma were estimated by FoundationOne CDx (F1CDx) as previously described (11,12).
The AF/MSAF status was categorized into two groups according to defined cutoff point with patients below and above the defined point; and the dividing points between groups were set from <1% to <50%. The prognostic value of AF/MSAF ratio that was below a defined cutoff point in overall survival (OS) was evaluated using Cox-proportional hazards regression. To find out the optimal prognostic cut-point for AF/MSAF, a series of AF/MSAF cutoffs were analyzed and the Wald-statistics of hazard ratios (HR) were fitted with a LOESS smoother (13). Then the structural break point of AF/MSAF was determined by Chow test (14,15), and was applied for separating patients into AFH present and AFH absent groups. The associations between presence of AFH and tumor size (SLD, sum of longest diameter of target lesions), or tumor-mutational burden (TMB) were also analyzed.
An independent cohort of 259 patients with advanced NSCLC receiving first-line EGFR-TKIs from the First Affiliated Hospital of Guangzhou Medical University (Jan 2012 to Dec 2018) was used for exploratory analysis to explore whether the prognostic role of AFH could be extended to different subsets of advanced NSCLC; genetic analysis of baseline plasma-derived ctDNA was confirmed by an targeted next generation sequencing panel (5) (Burning Rock Biotech, China) similar to the F1CDx test; ethical approval was obtained from institutional review boards and informed consent was waived due to the retrospective nature of this study. R software (Version 3.5.1) was used for all analyses and statistical significance was defined by a two-sided P<0.05.
Results
Development and analysis of allele frequency heterogeneity
This study used POPLAR and OAK trials of patients as primary cohort. A total of 853 advanced NSCLC patients that made up the blood biomarker-evaluable population were included for this analysis, with 784 (91.9%) EGFR-wild type patients.
In the univariable Cox proportional hazards regression model, low AF/MSAF ratio was found to be significantly associated with OS (Figure 1A). The cut-off valves (AF/MSAF of <1% to <30%) with statistically significant prognostic trends were further examined in the multivariate analysis. In the multivariate Cox proportional hazards regression model that controlled all baseline factors (see more details in Figure 1), low AF/MSAF ratio (i.e., AF/MSAF <10%) was demonstrated as an independent prognostic predictor in patients with advanced NSCLC.
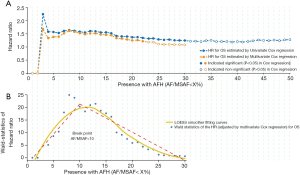
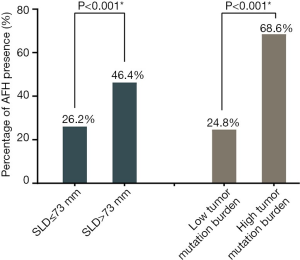
Figure 1B shows the LOESS fitting curves of Wald-statistics of OS-HR and AFH determined by different AF/MSAF cutoffs; and the corresponding structural break points for the AF/MSAF ratio. The structural break point analysis demonstrated that the presence of mutations with AF/MSAF <10% could yield the optimal predictive value to stratify patients with poor OS, which was applied for defining the presence of AFH. Results (Figure 2) showed that both large tumor size (46.4% vs. 26.2%) and high TMB (68.6% vs. 24.8%) were significantly associated with the presence of AFH.
Prognostic significance of allele frequency heterogeneity
In multivariate survival analyses of the primary cohort (POPLAR and OAK), independent from TMB and SLD, the presence of AFH (AF/MSAF <10%) significantly correlated with unfavorable OS in advanced NSCLC (HR: 1.52, 95% CI: 1.27–1.83, P<0.001) (Figure 3A); and median OS was shorter in AFH present group than in AFH absent group (7.7 vs. 11.7 months). Additionally, the presence of AFH was significantly associated with shorter OS regardless of treatment arms (Figure 3B,C) [immunotherapy (HR: 1.81, 95% CI: 1.38–2.37, P<0.001; median OS: 7.9 vs. 15.6 months) and chemotherapy (HR: 1.32, 95% CI: 1.01–1.71, P=0.039; median OS: 7.3 vs. 9.7 months), compared with AFH absent group].
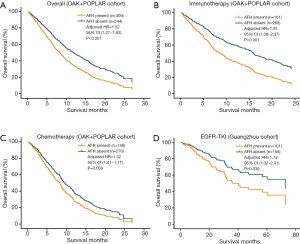
To unravel whether AFH defined by (AF/MSAF <10%) could identify patients with worse prognosis in different subsets of advanced NSCLC, another independent cohort of 259 patients with advanced NSCLC treated with EGFR-TKIs was used for exploratory analysis. In this exploratory cohort, results (Figure 3D) demonstrated that the presence of AFH was significantly associated with shorter OS (HR: 1.72, 95% CI: 1.02–2.91, P=0.039).
Discussion
Based on the impact of relatively low frequency mutations on survival, which is simultaneously determined by AF and MSAF, we determined the presence of AFH by the existence of certain mutations with AF/MSAF <10%. Low AF/MSAF ratio was found to be significantly associated with unfavorable survival after controlling tumor bulk and TMB, suggesting that AF/MSAF could be used as an independent factor for risk stratification of advanced NSCLC patients. Additionally, the presence of AFH (cases with mutation of AF/MSAF <10%) represented an adverse prognostic factor for OS across treatment arms (chemotherapy, immunotherapy or EGFR-TKIs).
The OAK&POPLAR cohort (predominantly EGFR-wild-type NSCLC) and EGFR-TKI cohort are two different subsets, they represent the main population in advanced NSCLC nowadays. The exploratory analysis further revealed poorer prognosis for AF/MSAF <10% in EGFR-TKI cohort, which gave important assumption that the AFH defined by AF/MSAF <10% might be a universal prognostic factor across different subsets of advanced NSCLC.
As an increasing proportion of patients undergo ctDNA profiling to search for targeted drugs or to calculate TMB during the course of diagnosis or treatment. Importantly, ctDNA has proven to be a potentially source for tumor burden monitoring and treatment outcome monitoring (16-20). This study initially links relatively low frequency mutations (AFH) to prognostic outcomes by using accessible indicators (AF/MSAF). Low AF/MSAF ratio provides a way to mirror the presence of AFH and hence to identify patients with poor outcomes. It is difficult to obtain comprehensive evaluation information of tumor heterogeneity in current clinical practice, however, the presence of the AFH defined by low AF/MSAF ratio thus provides an easily-accessible biomarker for poor prognosis that will allow for improved risk stratification in patients with advanced NSCLC.
There are several limitations to this study. First, the clinical investigation was retrospective and the detection of ctDNA might be challenged by the technical approach. Second, we cannot report the mechanism responsible for the prognostic role of low AF/MSAF ratio. Third, there remains a lack of validation on the optimal cut point for AFH definition, though, it should be noted that, this is the first study to present real-world data concerning AFH defined by ctDNA and prognosis in advanced NSCLC; additionally, the significance of this study is to illustrate that the prognostic impact of AFH defined by relatively low frequency mutations might be a general character in all subsets of advanced NSCLC, rather than finding out an absolute definition for AFH. Finally, the AFH merits further validation and improvement.
In conclusion, presence of AFH (AF/MSAF <10%) is an independent predictive factor for poor prognosis in advanced NSCLC. By adding this simple calculation to ctDNA profiling, which incurs no further cost, we can evaluate or monitor tumor heterogeneity and its prognosis. Further experimental evaluation is still warranted to verify these findings.
Acknowledgments
We sincerely thank Prof. Tony Mok and Roche China Lung Cancer Club for supporting this study.
Funding: This work was supported by the following funding: China National Science Foundation (Grant No. 81871893 & No. 81501996); Key Project of Guangzhou Scientific Research Project (Grant No. 201804020030).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Ethical approval was obtained from institutional review boards and informed consent was waived due to the retrospective nature of this study.
References
- McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016;351:1463-9. [Crossref] [PubMed]
- Wolf Y, Bartok O, Patkar S, et al. UVB-Induced Tumor Heterogeneity Diminishes Immune Response in Melanoma. Cell 2019;179:219-235.e21. [Crossref] [PubMed]
- Loeb LA. Human Cancers Express a Mutator Phenotype: Hypothesis, Origin, and Consequences. Cancer Res 2016;76:2057-9. [Crossref] [PubMed]
- Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 2013;497:108-12. [Crossref] [PubMed]
- Wang Z, Cheng Y, An T, et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): a phase 2, single-arm, multicentre clinical trial. Lancet Respir Med 2018;6:681-90. [Crossref] [PubMed]
- Yang XW, Xu W, Tian XY, et al. Diagnostic and prognostic value of KRAS mutations in circulating pancreatic ductal adenocarcinoma tumor DNA. Transl Cancer Res 2018;7:622-33. [Crossref]
- Rathod A, Hopkins A, Rowland A, et al. Circulating cell free deoxyribonucleic acid for tracking early treatment response and disease progression in advanced cancers. Transl Cancer Res 2017;6:S1530-40. [Crossref]
- Williams MJ, Werner B, Barnes CP, et al. Identification of neutral tumor evolution across cancer types. Nat Genet 2016;48:238-44. [Crossref] [PubMed]
- Fehrenbacher L, Spira A, Ballinger M, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 2016;387:1837-46. [Crossref] [PubMed]
- Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017;389:255-65. [Crossref] [PubMed]
- Gandara DR, Paul SM, Kowanetz M, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 2018;24:1441-8. [Crossref] [PubMed]
- Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [Crossref] [PubMed]
- Cleveland WS, Grosse E, Shyu WM. Chapter 8: Local regression models. In: Chambers JM, Hastie TJ. editors. Statistical Models in S. Wadsworth & Brooks/Cole Advanced Books & Software, 1992.
- Liang W, He J, Shen Y, et al. Impact of Examined Lymph Node Count on Precise Staging and Long-Term Survival of Resected Non-Small-Cell Lung Cancer: A Population Study of the US SEER Database and a Chinese Multi-Institutional Registry. J Clin Oncol 2017;35:1162-70. [Crossref] [PubMed]
- Zeileis A, Kleiber C, Krämer W, et al. Testing and dating of structural changes in practice. Comput Stat Data Anal 2003;44:109-23. [Crossref]
- Dawson SJ, Tsui DW, Murtaza M, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013;368:1199-209. [Crossref] [PubMed]
- Imamura F, Uchida J, Kukita Y, et al. Monitoring of treatment responses and clonal evolution of tumor cells by circulating tumor DNA of heterogeneous mutant EGFR genes in lung cancer. Lung Cancer 2016;94:68-73. [Crossref] [PubMed]
- Garcia-Saenz JA, Ayllon P, Laig M, et al. Tumor burden monitoring using cell-free tumor DNA could be limited by tumor heterogeneity in advanced breast cancer and should be evaluated together with radiographic imaging. BMC Cancer 2017;17:210. [Crossref] [PubMed]
- Wang YX, Li L, Cohen JD, et al. Prognostic Potential of Circulating Tumor DNA Measurement in Postoperative Surveillance of Nonmetastatic Colorectal Cancer. JAMA Oncol 2019;5:1118-23. [Crossref] [PubMed]
- McDonald BR, Contente-Cuomo T, Sammut SJ, et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci Transl Med 2019. [Crossref] [PubMed]


