
Cyclin D1 expression as a potential prognostic factor in advanced KRAS-mutant non-small cell lung cancer
Introduction
Mutations of Kirsten rat sarcoma (KRAS) in non-small cell lung cancer (NSCLC) are frequently found approximately 20–30% of lung adenocarcinomas in Western countries including Europe and America (1,2) and are associated with smoking (3) and poor prognosis (4). Though commonly found, KRAS mutation remains an un-targetable oncogene. In East Asian countries, lung cancers have been reported to have lower prevalence of KRAS mutations than those in western countries. Part of the reasons includes the higher prevalence (approximately 40–55%) of epidermal growth factor receptor (EGFR) mutations which is associated with non-smoking (5,6). Mutations of KRAS in East Asian regions reported from Japan, China and Taiwan were at 8–10% (5,6). Similarly, in Thailand, EGFR mutations are the most common driver mutations in NSCLC (7); however, the frequency of KRAS mutations is less described.
Cyclin D1, encoded by CCND1, is a CDK4/6-dependent regulator of the G1-S checkpoint of cell cycle. Cyclin D1 overexpression has been reported in many types of human cancers including lung cancer (8). In NSCLC, the reported frequencies of cyclin D1 overexpression were 18–76% while the frequencies of CCND1 amplifications were lower at 5–20% (9). These findings suggest additional mechanisms of cyclin D1 overexpression beyond CCND1 amplifications, particularly an activation of mitogenic signaling pathways including RAS-MEK-ERK pathways. Therefore, we hypothesized that KRAS-mutant NSCLC would drive cyclin D1 overexpression. To investigate this possibility, we determined the levels of cyclin D1 expression in correlation with KRAS mutation status in NSCLC tissues. Prognostic roles of cyclin D1 expression and KRAS mutations in NSCLC were also investigated.
Methods
DNA specimens
Eligible patients (aged ≥18 years) were those diagnosed with NSCLC at the King Chulalongkorn Memorial Hospital between January 2015 and July 2017. Based on prior report of mutually exclusive between EGFR and KRAS mutation, we excluded tissue samples with known EGFR mutations or inadequate amount (less than 100 ng) or quality of DNA specimens. Clinicopathological characteristics including demographic data, smoking status and TNM staging according to the 7th edition AJCC which were retrospectively reviewed from medical records. The study was approved by the Institutional Review Board of Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand (No. 560/59).
EGFR and KRAS mutation testing
Tumor specimens for all patients were obtained either from diagnostic biopsy or surgical procedures. DNAs were extracted from formalin-fixed, paraffin-embedded (FFPE) tissues using QIAamp DNA FFPE tissue kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. EGFR mutation testing platform was Cobas®EGFR mutation test v2 (Roche Diagnostics GmbH, Berlin, Germany) which was used to detect EGFR mutations in exons 18, 19, 20 and 21.
KRAS mutations at codons 12, 13 and 61 were examined using Cobas®KRAS mutation kit according to the manufacturer’s instructions (CE-IVD, Roche Diagnostics, Pleasanton, CA, USA). PCR amplifications and automated real-time mutation detections were performed using a Cobas z 480 analyzer (Roche Diagnostics, USA).
Immunohistochemistry for cyclin D1
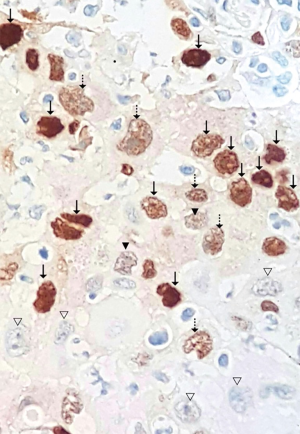
Two-micron FFPE sections were deparaffinized and rehydrated. Heat-induced epitope retrieval was performed using Dako PT link (Dako, Glostrup, Denmark). Immunostaining was performed using the automated staining systems, Dako Autostainer Link48 (Dako, Glostrup, Denmark). Primary antibody was FLEX monoclonal rabbit anti-human cyclin D1 clone EP12 ready-to-use (Dako, Glostrup, Denmark). The cyclin D1 immunostaining was evaluated by an experienced lung pathologist who was blinded from KRAS status. Cyclin D1 was evaluated for both its intensity and percentage of positivity. The intensity was classified into 0, 1+, 2+, 3+ where 0 was for no staining, 1+ for noticeable nuclear staining in 400× magnification, 2+ for distinct nuclear staining in 200× magnification, and 3+ for distinct nuclear staining in 100× magnification (Figure 1). The cyclin D1 immunoreactivity was previously reported as percentage of distinct nuclear staining (10) (defined as 2+ and 3+ in this study). Percentage of cyclin D1 expression was calculated using the following formula, % of cyclinD1 expression = (% cells with staining 2+) + (% cells with staining 3+).
Statistical analysis
The relationship between KRAS mutations and other clinicopathologic characteristics was analyzed using Chi-squared test or Fisher’s exact test (when the minimum expected count was less than 5). Binary logistic regression was performed to calculate odds ratio. Overall survival (OS) rate was measured from the date of diagnosis until the date of death from any causes and was analyzed by using the Kaplan-Meier method. Comparisons were done by using the log-rank test. The Cox proportional hazard model for survival was used for univariate and multivariate analyses. Median follow-up time was calculated using the reverse Kaplan-Meier method with the cut-off date on 28 September 2018. SPSS Statistical software version 21 (IBM Corp., Armonk, USA) was used to analyze the data. All tests were two-tailed and a P value <0.05 was considered statistically significance.
Results
Prevalence, clinical characteristics and prognosis of KRAS mutations
A total of 471 NSCLC patients received treatment at the King Chulalongkorn Memorial Hospital from January 2015 to July 2017. Of these patients, 262 (55.6%) had EGFR-mutant NSCLC. In NSCLC, EGFR and KRAS mutations are mutually exclusive (5), we thus identified KRAS mutations in NSCLC patients with EGFR wild-type (EGFR WT). After excluding 114 inadequate DNA specimens, there were 95 patients included for KRAS mutation testing (Figure 2). The median age was 67, and men accounted for 66.3% of the study population. Fifty-four point seven percent of the patients had history of smoking (Table 1). The majority of their histology (90.5%) was adenocarcinoma and 73.7% of the patients presented as stage IV at diagnosis. KRAS mutations were identified in 28 out of 95 patients with EGFR WT. The finding represented KRAS mutation rate of 7.8% of informative DNA status of the total population (28 out of 357, in which 262 were EGFR-mutant and 95 were EGFR WT NSCLC patients). Of 28 KRAS-mutant NSCLC patients, there were 26 (93%) patients with codon 12/13 mutations and 2 (7%) with codon 61 mutations.
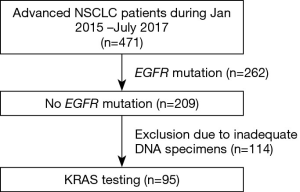
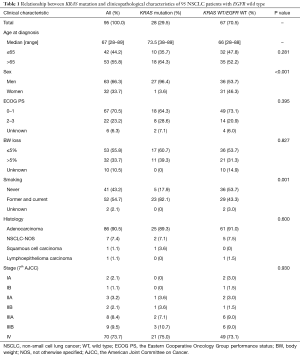
Full table
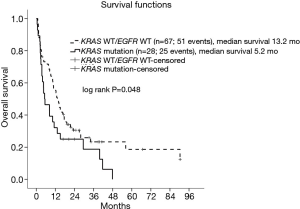
The clinical parameters that were significantly associated with KRAS mutations were sex (P<0.001) and smoking history (P=0.001) (Table 1). KRAS mutations were more frequently found in men [odds ratio 23.25; 95% confidence interval (CI): 2.98–181.13; P=0.003] and former/current smoker patients (odds ratio 5.71; 95% CI: 1.93–16.88; P=0.002). The median follow-up time of this study was 38.8 months and 80.0% (76 out of 95) of the patients died. The median OS of KRAS-mutant vs. KRAS WT/EGFR WT NSCLC patients were 5.2 vs. 13.2 months, respectively (Figure 3). KRAS-mutant NSCLC patients had statistically significant shorter OS (log rank P=0.05). Of note, patients with KRAS codon 61 mutations had limited survival comparing to those with KRAS codon 12/13 mutations (OS of 2.4 vs. 5.4 months).
Levels of cyclin D1 in KRAS mutant vs. KRAS WT/EGFR WT lung cancer
To test the hypothesis that KRAS-mutant NSCLC could drive cyclin D1 overexpression, immunohistochemistry of cyclin D1 was performed in 24 patients with KRAS mutant lung cancer and 26 patients with KRAS WT/EGFR WT lung cancer. The results demonstrated that cyclin D1 was mainly expressed in nucleus. The mean percentages of cyclin D1 expression were 68.83 (SD 27.11) and 50.50 (SD 27.5) in KRAS-mutant and KRAS WT/EGFR WT lung cancer, respectively. Cyclin D1 expression in KRAS-mutant lung cancer was significantly higher than that in KRAS WT/EGFR WT lung cancer (P=0.02) (Figure 4A).
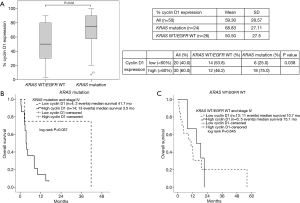
Prognosis role of cyclin D1 in KRAS mutant lung cancer
To investigate the prognostic role of cyclin D1, the expression levels of cyclin D1 were divided into two groups: high cyclin D1 and low cyclin D1 levels using the mean value of 60% as a cut point. High cyclin D1 levels were defined as the percentage of cyclin D1 expression of more than or equal 60% whereas low cyclin D1 levels were defined as cyclin D1 expression of less than 60%. Kaplan-Meier survival analyses of stage IV patients were performed to determine the prognostic role of cyclin D1 in KRAS-mutant and KRAS WT/EGFR WT NSCLC, separately.
In the KRAS-mutant NSCLC (n=18), patients with low cyclin D1 levels (n=4) had significantly better prognosis when compared with those with high cyclin D1 levels (n=14) (OS 41.7 vs. 3.5 months, P=0.037) (Figure 4B). In contrast, in the KRAS WT/EGFR WT NSCLC (n=19), there was no survival difference between patients with low (n=13) and high cyclin D1 (n=6) levels (OS 10.7 vs. 15.1 months, P=0.645) (Figure 4C).
Discussion
In the present study, we found a moderate prevalence of KRAS mutation, 7.8%, in Thai lung cancer. Similar to those of East Asian countries (11-13), where share a higher number of EGFR mutation especially in never or light smoking, KRAS mutation seems to be lower than those in Caucasian, 8–10% vs. 26.1% (5). However, we found that the prevalence of the underlying etiology for this group of KRAS mutation, smoking, was similar in both ethnicities. These results are consistent with the idea of a strong correlation between KRAS-mutant lung cancers to smoking. Consistent with previous studies (11,14,15), the prognosis of KRAS-mutant NSCLC was worse than that of KRAS WT/EGFR WT NSCLC (median survival of 5.2 vs. 13.2 months, P=0.048). NSCLC patients with KRAS codon 61 mutations had worse prognosis than those with KRAS codon 12/13 mutations. Different KRAS mutations subtypes have previously demonstrated different prognosis outcomes (16). In agreement with our results, KRAS codon 61 mutant lung cancers carried worse disease-free survival compared with those with KRAS codon 12 mutation (16). The worse outcome of KRAS codon 61 mutations could be explained by the more severely deficient in GTPase activity and relatively increased KRAS activity when compared with KRAS codon 12 or 13 mutations from an in-vitro study (17). The prevalence of KRAS mutations at codon 61 seems to be under-recognized as most studies selected only exon 2 (not exon 3) for determination of KRAS mutations. However, due to the small number of patients in this study, this needs validation in a larger patient population.
To our knowledge, this is the first study comparing the expression of cyclin D1 between NSCLC patients with KRAS mutation and those with KRAS WT/EGFR WT. Cyclin D1 expression in KRAS-mutant NSCLC was significantly higher than that found in KRAS WT/EGFR WT NSCLC (P=0.02). This could imply that KRAS-mutant NSCLC could drive cyclin D1 overexpression beyond CCND1 amplification via an activation of mitogenic signaling pathways including RAS-MEK-ERK pathways (17). In line with our findings, there was a small study exploring KRAS mutation and cyclin D1 mRNA expression in 30 patients with early stage (I–III) NSCLC comprising 9 adenocarcinomas and 21 squamous cell carcinomas. It was found that KRAS mutations was associated with cyclin D1 mRNA expression (18). In addition, KRAS mutations and CCND1 amplifications were found to be mutually exclusive according to Pan-Lung cancer database (TCGA, Nature Genetics 2016) (19-21).
Interestingly, among NSCLC patients with KRAS mutations, those with low cyclin D1 levels had much better prognosis than those with high cyclin D1 levels (OS 41.7 vs. 3.5 months) and numerically better than those with KRAS WT/EGFR WT (OS 13.2 months). We speculated that defective cyclin D1 expression downstream of MAPK pathway could render KRAS mutant tumor cells less aggressive. This finding should be interpreted with caution because of the low number of patients with KRAS-mutant NSCLC in this retrospective, single-center study. Despite the limitations of this study, this is the first study demonstrated the potential prognosis role of cyclin D1 in KRAS-mutant NSCLC patients that could be worth further exploring in larger size prospective studies.
Conclusions
In Thailand, there was a moderate prevalence of KRAS mutation in lung cancers at about 7.8%. Smoking was the strong clinical parameter corelated with KRAS mutations. Most KRAS-mutant NSCLC had high cyclin D1 expression and conferred poor prognosis. In contrast, KRAS-mutant NSCLC with low cyclin D1 level had much better prognosis.
Acknowledgments
Funding: This research was supported by grant to S Luangdilok for development of new faculty staff, Ratchadaphiseksomphot endowment fund for KRAS testing and Ratchadaphiseksomphot fund, Faculty of Medicine, Chulalongkorn University (RA60/114) for cyclin D1 expression.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Institutional Review Board of Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand (No. 560/59).
References
- Aviel-Ronen S, Blackhall FH, Shepherd FA, et al. K-ras mutations in non-small-cell lung carcinoma: a review. Clin Lung Cancer 2006;8:30-8. [Crossref] [PubMed]
- Ferrer I, Zugazagoitia J, Herbertz S, et al. KRAS-Mutant non-small cell lung cancer: From biology to therapy. Lung Cancer 2018;124:53-64. [Crossref] [PubMed]
- Slebos RJ, Hruban RH, Dalesio O, et al. Relationship between K-ras oncogene activation and smoking in adenocarcinoma of the human lung. J Natl Cancer Inst 1991;83:1024-7. [Crossref] [PubMed]
- Mitsudomi T, Steinberg SM, Oie HK, et al. ras gene mutations in non-small cell lung cancers are associated with shortened survival irrespective of treatment intent. Cancer Res 1991;51:4999-5002. [PubMed]
- Dearden S, Stevens J, Wu YL, et al. Mutation incidence and coincidence in non small-cell lung cancer: meta-analyses by ethnicity and histology (mutMap). Ann Oncol 2013;24:2371-6. [Crossref] [PubMed]
- Kohno T, Nakaoku T, Tsuta K, et al. Beyond ALK-RET, ROS1 and other oncogene fusions in lung cancer. Transl Lung Cancer Res 2015;4:156-64. [PubMed]
- Sriuranpong V, Chantranuwat C, Huapai N, et al. High frequency of mutation of epidermal growth factor receptor in lung adenocarcinoma in Thailand. Cancer Lett 2006;239:292-7. [Crossref] [PubMed]
- Musgrove EA, Caldon CE, Barraclough J, et al. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer 2011;11:558-72. [Crossref] [PubMed]
- Gautschi O, Ratschiller D, Gugger M, et al. Cyclin D1 in non-small cell lung cancer: a key driver of malignant transformation. Lung Cancer 2007;55:1-14. [Crossref] [PubMed]
- Keum JS, Kong G, Yang SC, et al. Cyclin D1 overexpression is an indicator of poor prognosis in resectable non-small cell lung cancer. Br J Cancer 1999;81:127-32. [Crossref] [PubMed]
- Sun JM, Hwang DW, Ahn JS, et al. Prognostic and predictive value of KRAS mutations in advanced non-small cell lung cancer. PLoS One 2013;8:e64816. [Crossref] [PubMed]
- Kosaka T, Yatabe Y, Onozato R, et al. Prognostic implication of EGFR, KRAS, and TP53 gene mutations in a large cohort of Japanese patients with surgically treated lung adenocarcinoma. J Thorac Oncol 2009;4:22-9. [Crossref] [PubMed]
- Xu J, He J, Yang H, et al. Somatic mutation analysis of EGFR, KRAS, BRAF and PIK3CA in 861 patients with non-small cell lung cancer. Cancer Biomark 2011-2012;10:63-9. [Crossref] [PubMed]
- Metro G, Chiari R, Bennati C, et al. Clinical outcome with platinum-based chemotherapy in patients with advanced nonsquamous EGFR wild-type non-small-cell lung cancer segregated according to KRAS mutation status. Clin Lung Cancer 2014;15:86-92. [Crossref] [PubMed]
- Nadal E, Chen G, Prensner JR, et al. KRAS-G12C mutation is associated with poor outcome in surgically resected lung adenocarcinoma. J Thorac Oncol 2014;9:1513-22. [Crossref] [PubMed]
- Izar B, Zhou H, Heist RS, et al. The prognostic impact of KRAS, its codon and amino acid specific mutations, on survival in resected stage I lung adenocarcinoma. J Thorac Oncol 2014;9:1363-9. [Crossref] [PubMed]
- Stephen AG, Esposito D, Bagni RK, et al. Dragging ras back in the ring. Cancer Cell 2014;25:272-81. [Crossref] [PubMed]
- Dragoj M, Milosevic Z, Bankovic J, et al. Association of CCND1 overexpression with KRAS and PTEN alterations in specific subtypes of non-small cell lung carcinoma and its influence on patients' outcome. Tumour Biol 2015;36:8773-80. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607-16. [Crossref] [PubMed]


