
SKA1/2/3 serves as a biomarker for poor prognosis in human lung adenocarcinoma
Introduction
Lung cancer was the most common malignancy with the highest mortality rate in the world (1,2). However, non-small cell lung cancer (NSCLC), accounting for about 85% of lung cancer, was a common pathological type of primary lung cancer, including large cell carcinoma, lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) (2-4). Among these types, LUAD accounted for about 40% of the incidence of lung cancer (4). In recent years, new targeted therapies had improved the survival rate of patients with LUAD. However, the prognosis of LUAD patients was still unsatisfactory, and the 5-year survival rate was still less than 25% (5). Therefore, it was urgent to identify more potential molecular targets in the diagnosis and treatment of LUAD to improve patient prognosis.
Spindle and kinetochore associated complex subunit (SKA) was composed of three members, namely SKA1/2/3, which stabilized spindle microtubules attaching to kinetochore (KT) in the middle stage of mitosis, thus ensuring the accomplishment of mitosis process (6-9). Studies had found that SKA1/2/3 were dysregulated and closely related to prognosis in liver cancer, breast cancer, cervical cancer and other malignant tumors (10-12). In hepatocellular carcinoma (HCC) tissues, SKA1 was significantly upregulated, and correlated with alpha-fetoprotein (AFP), tumor size and TNM stage. Liver cancer patients with high SKA1 expression had a worse prognosis than those with low SKA1 expression (8). The expression of SKA2 in breast cancer was also elevated, and the prognosis of breast cancer patients with high SKA2 was poor. Interference with SKA2 inhibited the proliferation, migration and invasion of breast cancer cells (9). SKA3 level was increased in cervical cancer, and patients with high SKA3 have a poor prognosis. Overexpression of SKA3 promoted proliferation and migration of cervical cancer cells and accelerated tumor growth (10). Therefore, SKA1/2/3 may serve as a potential biomarker to predict tumor prognosis. Current studies had found that SKA1 in NSCLC was upregulated, and related to tumor type, clinical stage and lymph node metastasis, and high SKA1 promoted proliferation, metastasis and cisplatin resistance of NSCLC cells (11). Nevertheless, the potential clinical value of SKA1/2/3, especially in terms of prognosis and development of NSCLC, had not been fully elucidated.
The development of microarray and RNA sequencing technology made RNA research an essential part of biomedical research. Thus, in this study, on the basis of various databases, we analyzed the expression of SKA members, and explored its correlation with clinicopathological features and prognosis, and its potential regulatory mechanism in LUAD patients.
Methods
Clinical samples
Tumor and adjacent non-tumorous tissues of 30 patients with LUAD, who underwent surgical treatment in the Department of Thoracic Surgery, Affiliated Hospital of Zunyi Medical University from May 2018 to April 2019, were collected. This study was approved by the Ethics Committee of the Affiliated Hospital of Zunyi Medical University. All 30 patients with LUAD signed written informed consent and did not receive other special treatment before surgery.
Analysis of Oncomine
Oncomine (www.oncomine) gene expression microarray database was an accessible online microarray database that helped us understand genome-wide expression related research. Oncomine was used to analyze RNA levels of SKA1/2/3 in LUAD. Screening criteria: P<0.0001; Multiple change =1.5; The data type was mRNA; Analysis type: LUAD vs. normal tissue.
Analysis of UALCAN and GEPIA
UALCAN (ualcan.path.uab.edu) and GEPIA were open databases for in-depth analysis of gene expression data of TCGA, and were applied to analyze the expression of SKA1/2/3 and its Hub gene in LUAD and normal samples, the correlation between SKA1/2/3 and the clinicopathological characteristics of LUAD (cancer stage, tumor grade, race, weight or stage, etc.) and its potential prognostic value.
Analysis of CbioPortal
CBioPortal database (www.cbioportal.org) integrated data from multiple databases including the cancer genome atlas (TCGA) and the International Cancer Genome Gonsortium (ICGC), providing cancer genome data, comprehensive analysis of genome data and clinical data. MRNA levels of SKA member (RNA Seq V2 RSEM) was analyzed by cBioPortal database.
Biological functional analysis
Gene expression data of LUAD in HTSeq-FPKM were downloaded from TCGA official website for analysis, and gene expression data of 535 patients were analyzed. The co-expressed genes of SKA1/2/3 were screened with Pearson correlation coefficients (|r| >0.4 and P<0.001). Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genome (KEGG) analysis were performed on co-expressed genes with the package R “clusterProfiler” to explore possible biological functions and signaling pathways affected by SKA1/2/3 (13). GO analysis included biological process (BP), cell composition (CC) and molecular function (MF) (P<0.05 was statistically significant). TCGA gene data set was analyzed by GSEA (14). A protein-protein interaction (PPI) network of SKA co-expressed genes were constructed using a STRING (string-db.org) database, and combined score of >0.7 was considered statistically significant (15). The obtained PPI network was imported into Cytoscape 3.6.1 software, and the first 10 genes were defined as hub genes using CytoHubba plug-in as the hub gene screening standard (16), and relevant analysis was conducted.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA of tissues were collected according to the instructions of Trizol kit (Invitrogen), and cDNA was synthesized with the reverse transcription kit (Takara). The expression of SKA1, SKA2 and SKA3 was detected by qRT-PCR. GAPDH was used as internal control. Primer sequences: SKA1 forward, 5'-CCTGAACCCGTA AAGAAGCCT-3'; SKA1 reverse, 5'-TCATGTACGAAGGAACACCATTG-3'; SKA2 forward, GCCGCATTTGTGCTACTGTG; SKA2 reverse, CTCTGCCGCAGTTTT CTCTT; SKA3 forward, TGAGCGGTACATCGTATCCCA; SKA3 reverse, GGGG TTACAATTACGGGCTCT; GAPDH forward, 5'-AACGGATTTGGTCGTATTG-3'; GAPDH reverse, 5'-GGAAGATGGTGATGGATT-3'.
Statistical analysis
SPSS 20.0 software was used for statistical analysis, and GraphPad Prism 5 software was used for graphing. Normally distributed measurement data are expressed as mean ± standard deviation (
Results
The expression of SKA1/2/3 in LUAD was elevated
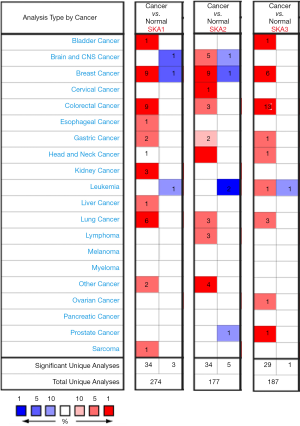
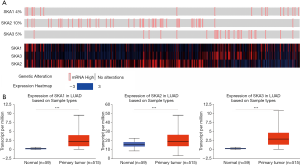
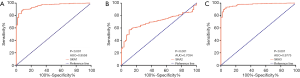
Oncomine database was used to compare the mRNA levels of SKA1/2/3 between LUAD and normal tissues. It showed that mRNA levels of SKA1, SKA2 and SKA3 were up-regulated in breast, rectal, gastric and lung cancer patients (Figure 1). Further analysis showed that SKA1 was overexpressed in LUAD patients in Garber database, with fold change of 2.379 and P value of 8.50E-9. In the Su database, SKA1 was also overexpressed in LUAD, with fold change of 2.633 and P value of 6.55E-5. In the Hou data set, mRNA levels of SKA2 and SKA3 were significantly increased in patients with LUAD, with fold change of 1.756 and 1.601, and P values of 3.61E-9 and 6.12E-12, respectively (Table 1). In addition, by means of UALCAN, CbioPortal and TCGA database, it’s found that the change rates of SKA1/2/3 mRNA in LUAD was 4%, 10% and 5%, respectively (Figure 2A), and SKA1/2/3 mRNA level was elevated in LUAD (Figure 2B), and the AUC was 0.9558, 0.7034 and 0.9775, respectively (P<0.05) (Figure 3).

Full table
High SKA1/2/3 was closely related to smoking, tissue subtypes and prognosis of LUAD patients
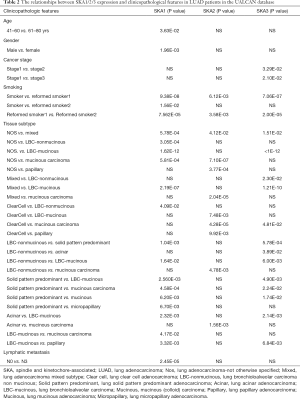
In the UALCAN database, high SKA1/2/3 was correlated with clinicopathological features and prognosis of LUAD patients. High SKA1 was related to age, gender, smoking, lymph node metastasis and tissue subtypes. High SKA2 was significantly associated with smoking and tissue subtypes. High SKA3 was significantly linked to smoking, clinical stage, lymph node metastasis and tissue subtypes (P<0.05, Table 2). In addition, LUAD patients with high SKA1/2/3 expressions had a worse prognosis than those with low expression (P<0.01, Figure 4).
Full table

Co-expressed genes of SKA1/2/3
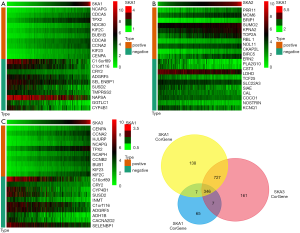
In TCGA transcriptome database, there were 875 positively correlated genes and 343 negatively correlated genes of SKA1, 390 positively correlated genes and 35 negatively correlated genes for SKA2, and 821 positive and 420 negative genes for SKA3 (http://cdn.amegroups.cn/static/application/0236bc8b212a426d6e7e5678d30c87b4/tlcr.2020.01.20-1.pdf). The top 10 co-expressed genes with positive and negative correlation of SKA1/2/3 were shown in the form of heat map (Figure 5A,B,C). In addition, Venn diagram indicated that there were 346 intersections of SKA1/2/3 co-expressing genes (Figure 5D and http://cdn.amegroups.cn/static/application/18e59453f127731692b51a9c969cea01/tlcr.2020.01.20-2.pdf).
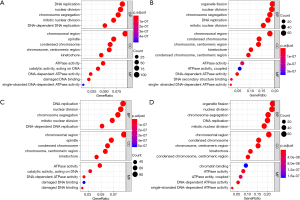
GO and KEGG analysis
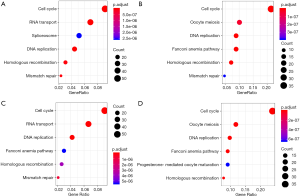
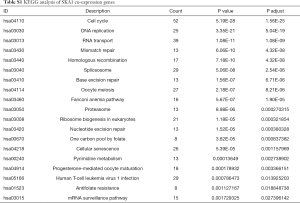
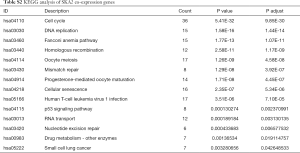
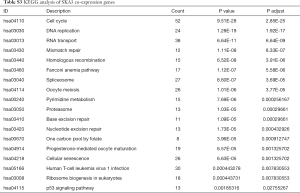
To further understand the potential role of SKA1/2/3 in LUAD, GO and KEGG analyses was performed on SKA1/2/3 co-expressed genes. Results revealed that SKA1/2/3 co-expressed genes were mainly involved in organelle division, DNA replication and mitosis (Figure 6 and http://cdn.amegroups.cn/static/application/578ef33e6dd8ccb644a9d5db89d11d50/tlcr.2020.01.20-3.pdf). KEGG pathway analysis displayed that SKA1/2/3 co-expressed genes were enriched in regulating cell cycle, DNA replication and p53 signaling pathway (Figure 7 and Tables S1-S4). These results suggested that SKA1/2/3 worked by participating in cell cycle, DNA replication and p53 signaling pathway in LUAD.
Full table
Full table
Full table

Full table
Analysis of Hub genes through PPI network
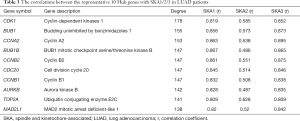
PPI network analysis showed the representative 10 Hub genes were CDK1, BUB1, CCNA2, BUB1B, CCNB2, CDC20, CCNB1, AURKB, TOP2A and MAD2L1 (Table 3). GEPIA analysis demonstrated that CDK1, BUB1, CCNA2, CDC20, CCNB2, CCNB1, BUB1B, AURKB, TOP2A and MAD2L1 were increased in LUAD (P<0.05) (Figure 8), and related to the overall survival (OS) of LUAD patients (Figure S1). Additionally, CDK1, BUB1, CCNB1, CCNB2, BUB1B and AURKB were significantly related to the disease-free progression (DFS) of LUAD patients (Figure S2).
Full table
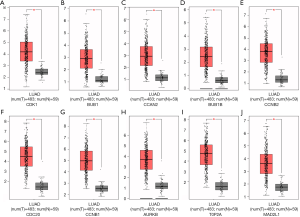
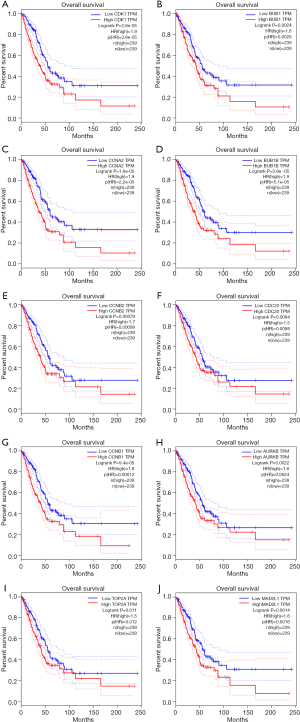
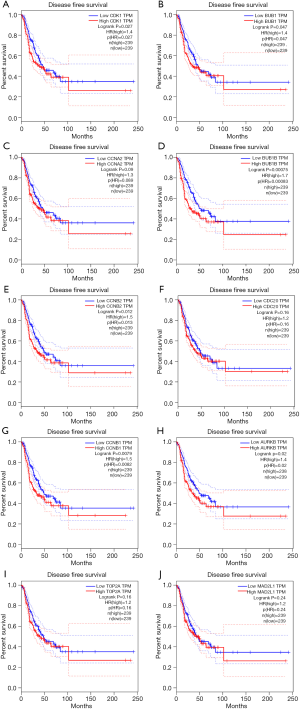
Verification of SKA1/2/3 mRNA expression in LUAD tissue samples
The mRNA of SKA1/2/3 in 30 human LUAD and non-cancerous tissues were detected by qRT-PCR, and the correlation between their expressions with clinical pathological features were analyzed (Table 4). Among them, SKA1 expression was increased in 22 of the 30 LUAD tissues (P<0.05). The mRNA levels of SKA2 and SKA3 were increased in 18 LUAD tissues, but there were no statistical significance. The mRNA levels of SKA1 was significantly correlated with the gender and smoking history, that of SKA2 was significantly related to the age, and that of SKA3 was significantly associated with smoking history and lymph node metastasis in LUAD patients.
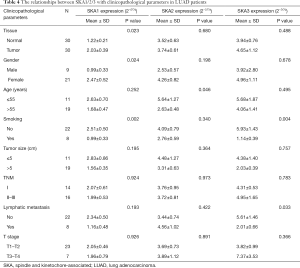
Full table
Discussion
Several studies had shown that SKA1/2/3 were not only involved in mitosis, but also in apoptosis and tumor development. Abnormal expression or activation of SKA1/2/3 was a common phenomenon in malignancies, and it’s demonstrated that SKA was significantly associated with malignancy. SKA1 was upregulated in salivary adenoid cystic carcinoma (SACC), and SKA1 overexpression was positively correlated with advanced SACC and its histological morphology, tumor recurrence, nerve invasion and survival time (16). Knockdown of SKA1 inhibited SACC progress (17). SKA1 expression was elevated in NSCLC, and high SKA1 expression contributed to the tumor progression and increasing of malignancy. Increased expression of SKA1 also promoted proliferation and metastasis of NSCLC cells. In addition, SKA1 leaded to cisplatin resistance in NSCLC cells and inhibited tumor cell apoptosis through ERK1/2 and AKT-mediated signaling pathways (18).
SKA was an essential component of motor-binding microtubules. Abnormal expression of SKA caused defects in spindle detection points, and it played a critical role in cell cycle regulation and tumor occurrence and development. SKA1 expression was elevated in salivary adenoid cystic carcinoma (SACC), and SKA1 overexpression was positively correlated with advanced SACC and its histological morphology, tumor recurrence, nerve invasion and survival time (17). Knockdown of SKA1 inhibited SACC progress (19). And SKA1 level was increased in NSCLC, and high SKA1 expression contributed to tumor progression. Increased expression of SKA1 promoted proliferation and metastasis of NSCLC cells. Furthermore, SKA1 leaded to cisplatin resistance in NSCLC cells and inhibited tumor cell apoptosis through ERK1/2 and AKT-mediated signaling pathways (18).
SKA2 was involved in mitosis and was crucial to the regulation of mitosis. In recent years, it’s found that SKA2 was up-regulated in a variety of human malignant tumors. SKA2 expression was elevated in breast cancer, and was associated with tumor stage and lymph node metastasis. SKA2 inhibited tumor cell migration and invasion in breast cancer cells, and the markers of epithelial mesenchymal transformation (EMT) such as fibronectin, N-cadherin, and vimentin (19), and the prognosis of breast cancer patients with high SKA2 expression was poor (12). In addition, up-regulation of SKA2 reversed the inhibitory effect of miR-520a-3p on proliferation, migration and invasion of gastric cancer cells and participated in the progression of gastric cancer (20).
SKA3, a key member of SKA, lies in the outer layer of complex’s centromere and plays an important role in mitosis (10,21-23). It was found that SKA3 expression was increased in colorectal cancer (CRC), and high SKA3 expression was associated with chromosomal instability (CIN). Knockout of SKA3 directly inhibited the growth and migration of CRC cells, induced CRC cell cycle arrest and apoptosis, and participated in the progression of CRC (24). Besides, increased SKA3 expression was significantly associated with OS in breast cancer patients (25), and down-regulated SKA3 inhibited the proliferation of breast cancer cells (26). Furthermore, SKA3 expression was enhanced in renal cell carcinoma tissues. Knockdown of SKA3 inhibited the growth and migration of renal cancer cells (27). SKA3 overexpression enhanced the proliferation and migration of cervical cancer cells by increasing the expression of p-AKT, cyclinE2, CDK2, cyclinD1, CDK4, E2F1 and p-Rb proteins in cervical cancer cells (10). Up-regulation of GNL3 and SKA3 reduced the migration and invasion ability of prostate cancer cells, suggesting that SKA3 was related to the progression of prostate cancer. In summary, SKA1/2/3 was associated with cell cycle, cell proliferation and apoptosis, and poor prognosis in cancer patients. However, the expression pattern and role of SKA1/2/3 in LUAD were unclear. In this study, we investigated the expression of SKA complex members, correlations with clinicopathological features and prognostic value, and potential functional mechanisms in LUAD.
In this study, we found that SKA1/2/3 was up-regulated, and the high expression of SKA1/2/3 was related to smoking and tissue subtypes in LUAD patients. In addition, increased SKA1/2/3 mRNA levels were associated with overall survival (OS). Therefore, they may serve as new targets for anticancer therapy. GO and KEGG analysis showed that SKA1/2/3 co-expressed genes were enriched in regulating cell cycle, DNA replication and p53 signaling pathway. Hub genes in PPI network were CDK1, BUB1, CCNA2, CDC20, CCNB2, CCNB1, BUB1B, AURKB, TOP2A and MAD2L1. GEPIA analysis indicated that Hub genes were highly expressed and negatively correlated with the prognosis of LUAD patients. High-throughput low-power laser irradiation (HF-LPLI) up-regulated survivin activity in human LUAD cells through reactive oxygen species (ROS)/cdc25c protein phosphatase (cdc25c)/CDK1 signaling pathway. Up-regulation of survivin activity reduced HF-LPLI-induced apoptosis of LUAD cells, while down-regulation of survivin activity promoted apoptosis of tumor cells (28). BUB1 was a mitotic checkpoint of serine/threonine kinase that acted as an oncogene in tumorigenesis and progression in various cancers, including cell proliferation, tumor growth, metastasis, and patient prognosis. Elevated BUB1 expression level was directly associated with poor prognosis of glioma patients. ShRNA silencing of BUB1 inhibited proliferation and tumorigenicity of glioblastoma tumor cells. Moreover, BUB1 silencing also promoted cytotoxic effects of irradiation on glioblastoma cells (29). CCDC106 increased the expression of CCNA2 and CCNB1, and then contributed to the proliferation of LUAD cells (30).
In addition, by collecting tissue samples from 30 patients with clinical LUAD and testing the levels of SKA1/2/3 mRNA, we found that the levels of SKA1/2/3 in LUAD tissues were all higher than those in adjacent tissues. Unfortunately, this result was not completely statistically significant. We analyzed the potential clinical value of SKA1/2/3 in the pathogenesis and development of LUAD and its related carcinogenic signaling pathways, so as to provide clues for multi-targeting and SKA1/2/3 mediated target therapy. However, it remained some limits in present study: (I) the clinical sample size was insufficient; (II) SKA member interactions and mechanisms regulating the development and progression of LUAD required additional clinical samples of LUAD; (III) further verification of SKA function at the cellular level was needed.
In general, SKA1/2/3 mRNA level in LUAD was significantly up-regulated, and was significantly correlated with smoking, tissue classification and prognosis of LUAD patients. These results suggested that SKA1/2/3 may serve as potential prognostic biomarkers and therapy targets for LUAD. This study may contribute to a better understanding of the molecular basis of LUAD and facilitate the development of SKA-mediated LUAD therapies. However, further experimental studies are still needed to confirm our findings and promote SKA’s clinical application as a prognostic indicator or therapeutic target for LUAD.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr.2020.01.20). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by the Ethics Committee of Affiliated Hospital of Zunyi Medical College (No. 2019102601).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Moravcikova E, Krepela E, Donnenberg VS, et al. BOK displays cell death-independent tumor suppressor activity in non-small-cell lung carcinoma. Int J Cancer 2017;141:2050-61. [Crossref] [PubMed]
- Weber M, McWilliams A, Canfell K. Prospects for cost-effective lung cancer screening using risk calculators. Transl Cancer Res 2019;8:S141-4. [Crossref]
- Chiba R, Morikawa N, Sera K, et al. Elemental and mutational analysis of lung tissue in lung adenocarcinoma patients. Transl Lung Cancer Res 2019;8:S224-34. [Crossref] [PubMed]
- Xia L, Zhu Y, Zhang C, et al. Decreased expression of EFCC1 and its prognostic value in lung adenocarcinoma. Ann Transl Med 2019;7:672. [Crossref] [PubMed]
- Macheleidt IF, Dalvi PS, Lim SY, et al. Preclinical studies reveal that LSD1 inhibition results in tumor growth arrest in lung adenocarcinoma independently of driver mutations. Mol Oncol 2018;12:1965-79. [Crossref] [PubMed]
- Auckland P, Clarke NI, Royle SJ, et al. Congressing kinetochores progressively load Ska complexes to prevent force-dependent detachment. J Cell Biol 2017;216:1623-39. [Crossref] [PubMed]
- Zhou L, Ouyang L, Chen K, et al. Research progress on KIF3B and related diseases. Ann Transl Med 2019;7:492. [Crossref] [PubMed]
- Sivakumar S, Gorbsky GJ. Phosphatase-regulated recruitment of the spindle- and kinetochore-associated (Ska) complex to kinetochores. Biology Open 2017;6:1672-9. [Crossref] [PubMed]
- Lange KI, Suleman A, Srayko M. Kinetochore Recruitment of the Spindle and Kinetochore-Associated (Ska) Complex Is Regulated by Centrosomal PP2A in. Genetics 2019;212:509-22. [Crossref] [PubMed]
- Hu R, Wang MQ, Niu WB, et al. SKA3 promotes cell proliferation and migration in cervical cancer by activating the PI3K/Akt signaling pathway. Cancer Cell Int 2018;18:183. [Crossref] [PubMed]
- Chen Y, Zhao J, Jiao Z, et al. SKA1 overexpression is associated with poor prognosis in hepatocellular carcinoma. BMC Cancer 2018;18:1240. [Crossref] [PubMed]
- Wang Y, Zhang C, Mai L, et al. PRR11 and SKA2 gene pair is overexpressed and regulated by p53 in breast cancer. BMB Rep 2019;52:157-62. [Crossref] [PubMed]
- Zhou Q, Hou Z, Zuo S, et al. LUCAT1 promotes colorectal cancer tumorigenesis by targeting the ribosomal protein L40-MDM2-p53 pathway through binding with UBA52. Cancer Sci 2019;110:1194-207. [Crossref] [PubMed]
- Lou Y, Yu Y, Xu X, et al. Long non-coding RNA LUCAT1 promotes tumourigenesis by inhibiting ANXA2 phosphorylation in hepatocellular carcinoma. J Cell Mol Med 2019;23:1873-84. [Crossref] [PubMed]
- Cheng S, Li X, Lin L, et al. Identification of Aberrantly Expressed Genes during Aging in Rat Nucleus Pulposus Cells. Stem Cells Int 2019;2019:2785207. [Crossref] [PubMed]
- Zhao L, Jiang L, Du P, et al. Expression of SKA1 and MMP-9 in primary salivary adenoid cystic carcinoma: Correlation with tumor progression and patient prognosis. Acta Otolaryngol 2016;136:575-9. [Crossref] [PubMed]
- Zhao LJ, Yang HL, Li KY, et al. Knockdown of SKA1 gene inhibits cell proliferation and metastasis in human adenoid cystic carcinoma. Biomed Pharmacother 2017;90:8-14. [Crossref] [PubMed]
- Shen L, Yang M, Lin Q, et al. SKA1 regulates the metastasis and cisplatin resistance of non-small cell lung cancer. Oncol Rep 2016;35:2561-8. [Crossref] [PubMed]
- Ren Z, Yang T, Zhang P, et al. SKA2 mediates invasion and metastasis in human breast cancer via EMT. Mol Med Rep 2019;19:515-23. [PubMed]
- Su H, Ren F, Jiang H, et al. Upregulation of microRNA-520a-3p inhibits the proliferation, migration and invasion via spindle and kinetochore associated 2 in gastric cancer. Oncol Lett 2019;18:3323-30. [PubMed]
- Gaitanos TN, Santamaria A, Jeyaprakash AA, et al. Stable kinetochore- microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J 2009;28:1442-52. [Crossref] [PubMed]
- Theis M, Slabicki M, Junqueira M, et al. Comparative profiling identifies C13orf3 as a component of the Ska complex required for mammalian cell division. EMBO J 2009;28:1453-65. [Crossref] [PubMed]
- Zhang QH, Qi ST, Wang ZB, et al. Localization and function of the Ska complex during mouse oocyte meiotic maturation. Cell Cycle 2012;11:909-16. [Crossref] [PubMed]
- Chuang TP, Wang JY, Jao SW, et al. Over-expression of AURKA, SKA3 and DSN1 contributes to colorectal adenoma to carcinoma progression. Oncotarget 2016;7:45803-18. [Crossref] [PubMed]
- Tang D, Zhao X, Zhang L, et al. Identification of hub genes to regulate breast cancer metastasis to brain by bioinformatics analyses. J Cell Biochem 2019;120:9522-31. [Crossref] [PubMed]
- Jiao X, Hooper SD, Djureinovic T, et al. Gene rearrangements in hormone receptor negative breast cancers revealed by mate pair sequencing. BMC Genomics 2013;14:165. [Crossref] [PubMed]
- Yamada Y, Arai T, Kojima S, et al. Anti-tumor roles of both strands of the duplex: their targets and are involved in the pathogenesis of renal cell carcinoma. Oncotarget 2018;9:26638-58. [Crossref] [PubMed]
- Chu J, Wu S, Xing D. Survivin mediates self-protection through ROS/cdc25c/CDK1 signaling pathway during tumor cell apoptosis induced by high fluence low-power laser irradiation. Cancer Lett 2010;297:207-19. [Crossref] [PubMed]
- Yu H, Zhang S, Ibrahim AN, et al. Serine/threonine kinase BUB1 promotes proliferation and radio-resistance in glioblastoma. Pathol Res Pract 2019;215:152508. [Crossref] [PubMed]
- Zhang X, Zheng Q, Wang C, et al. CCDC106 promotes non-small cell lung cancer cell proliferation. Oncotarget 2017;8:26662-70. [PubMed]


