EGFR mutation tracking predicts survival in advanced EGFR-mutated non-small cell lung cancer patients treated with osimertinib
Introduction
Osimertinib has been established as standard treatment for advanced epidermal growth factor receptor (EGFR) mutated non-small cell lung cancer (NSCLC). The superior efficacy of osimertinib has been shown in two phase III trials (1,2). In the AURA3 trial, osimertinib prolonged progression-free survival (PFS) over platinum-based chemotherapy in pretreated patients with advanced EGFR-mutated NSCLC and T790M-mediated resistance (1). The results of this trial led to the approval of second-line osimertinib, for patients with confirmed T790M mutation in tumor tissue or cell-free plasma DNA. In the FLAURA phase III trial, osimertinib showed superior efficacy compared to erlotinib and gefitinib in the first-line treatment of NSCLC with common EGFR mutations, irrespective of the T790M status (2). Osimertinib was therefore approved as a first-line treatment for advanced NSCLC with EGFR exon 19 deletions or L858R mutations.
The analysis of T790M in plasma-based circulating tumor DNA (ctDNA) complemented by tumor tissue biopsies in case of a T790M-negative result in plasma is currently considered the preferred strategy to select EGFR-mutated NSCLC patients for second-line therapy with osimertinib (3-8). Continuous monitoring of the tumor genotype could also be important for early identification of emerging changes in tumor biology that negatively affect treatment outcome. In particular, tracking of EGFR mutations may be important for response evaluation, real-time assessment of resistance evolution and treatment guidance (9-12). To this end, we investigated the clinical utility of EGFR mutation tracking in plasma ctDNA after start of osimertinib therapy in patients who developed resistance to prior treatment with EGFR tyrosine kinase inhibitors (TKIs).
Methods
Patients
Patients with metastatic EGFR-mutant NSCLC received second-line osimertinib after detection of a T790M mutation in plasma ctDNA and/or tissue re-biopsy at the Department of Respiratory and Critical Care Medicine, and Ludwig Boltzmann Institute of COPD and Respiratory Epidemiology, Otto Wagner Hospital, Vienna, between February 2016 and August 2017. Diagnostic biopsies were available from each included patient and showed adenocarcinoma histology and EGFR mutations in all cases. Blood sampling was performed as part of diagnostic routine procedures. EGFR mutation analyses were carried out at the Institute of Cancer Research, Department of Medicine I, Medical University of Vienna. The collection and analysis of blood samples was approved by the local ethics committee (EK No. 1132/2016) and informed consent was obtained from all patients. Forty patients had been included in a previous study (6).
Plasma genotyping
Preparation and storage of blood samples was done as previously described (6). In brief, Cell-Free DNA Blood Collection Tubes (Streck, La Vista, NE, USA) or Cell Free DNA Blood Collection Tubes (Roche, Pleasanton, CA, USA) were used for blood sampling and one blood sample (8 mL) was obtained from all patients at each time point.
For plasma isolation, blood samples were centrifuged at increasing speed (10 minutes at 200 g followed by 10 minutes at 1,600 g). The supernatant was collected and centrifuged again for 10 minutes at 1,900 g.
For ddPCR, we extracted ctDNA from 2 mL plasma using the QIAamp circulating nucleic acid kit (Qiagen, Venlo, The Netherlands) according to manufacturer’s instructions.
EGFR deletions in exon 19, L858R, L861Q, S768I, T790M and C797S mutations were assessed by using the QX-200TM ddPCR system (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions.
Custom assays for ddPCR from Life Technologies (Carlsbad, CA, USA) and ddPCR assays from Bio-Rad were used for EGFR mutation analysis as previously described (6). We used QuantaSoft analysis software (Bio-Rad) for qualitative and quantitative mutation analysis. All ddPCR assays were performed blinded to the study endpoint and analyzed in triplicate. Finally, the absolute copy-number of mutant alleles per mL of plasma was calculated. We used a threshold of >1 copy/mL for positivity of each mutation analyzed. Plasma ctDNA was termed positive if any EGFR mutation was detected.
Statistical analyses
We used PFS as assessed by investigators as the primary study endpoint. PFS was defined as the time from first osimertinib dose to disease progression or death from any cause, whichever came first. Overall survival (OS) and response rate (RR) were secondary endpoints. OS was defined as the time from first osimertinib dose to death from any cause. RR was defined as the percentage of patients with response (complete or partial) at restaging after osimertinib initiation. Regular CT scans of the chest and abdomen, usually performed every 6–8 weeks were used to assess tumor response at the medical center of the treating physician according to institutional practice. Additionally, response was confirmed post hoc using Response Evaluation Criteria in Solid Tumors (RECIST) 1.1.
Characteristics of patients included age, gender, presence or absence of extra-thoracic metastases, tissue genotype at diagnosis, and previous EGFR TKI therapy. We used the chi-square test or fisher’s exact test to assess associations of plasma genotyping results with clinical parameters and with treatment response. Survival probabilities were calculated with the product limit method according to Kaplan-Meier. Hazard ratios (HRs) and their confidence intervals (CIs) were estimated using univariable and multivariable Cox proportional hazards models. For the multivariable analyses we used full models and stepwise backward selection models that included age (as continuous variable), gender (male, female), presence or absence of extra-thoracic metastases (thoracic, extra-thoracic), tissue genotype at diagnosis (EGFR deletions in exon 19, L858R, other EGFR mutations), previous EGFR TKI therapy (afatinib, erlotinib, gefitinib, >1 EGFR TKI), and ctDNA status (positive, negative) or status of the activating EGFR mutation (detectable, not detectable). All reported P values are two sided. All analyses were performed using IBM SPSS Statistics software, version 25 (SPSS, IBM Corporation, Armonk, NY, USA).
Results
Plasma samples of 141 patients who progressed under first- or second-generation EGFR-TKI therapy were centrally tested for activating EGFR mutations (EGFR exon 19 deletions, L858R, L861Q, S768I) and the T790M mutation by ddPCR.
At the start of osimertinib, all 141 patients were T790M positive and 122 of 141 (87%) patients were also positive for the corresponding activating EGFR mutations. The 19 patients who were T790M positive but in whom the activating EGFR mutation was not detectable were also treated with osimertinib.
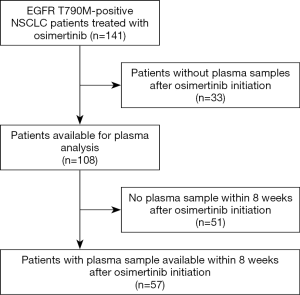
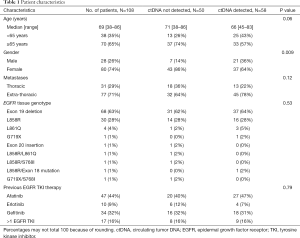
EGFR mutation tracking in plasma ctDNA was performed in 108 patients (including 15 of 19 T790M-positive but activating EGFR mutation-negative patients) undergoing osimertinib treatment for T790M-positive NSCLC after progression under treatment with an EGFR TKI and in whom a plasma sample was available. The study flowchart is shown in Figure 1. Blood sampling was performed at several time points during osimertinib treatment starting with the first osimertinib dose. The number of serial samples collected from each patient throughout osimertinib therapy to assess EGFR deletions in exon 19, L858R, L861Q, S768I, T790M, and C797S mutations varied between two and thirteen samples. As shown in Table 1, only patients with lung adenocarcinoma histology and stage IV disease were enrolled. Prior EGFR TKIs included gefitinib, erlotinib and afatinib and osimertinib was initiated as second-line treatment without exceptions. All patients were T790M mutation-positive assessed by plasma genotyping and/or tissue re-biopsy testing. The tissue genotype at diagnosis included EGFR exon 19 deletions in 68 (63%) patients, L858R in 30 (28%), and other EGFR mutations in 10 (9%) patients.
Full table
Fifty-eight out of 108 (54%) patients treated with osimertinib were ctDNA positive. Samples were classified as positive if any EGFR activating or resistance mutation was detected. ctDNA was more frequently detected in males than in females (P=0.009) but no other correlation between ctDNA status and clinical variables was seen (Table 1). Eighty-two percent of the patients responded to osimertinib.
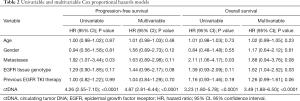
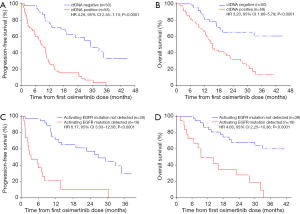
At a median follow-up of 32.3 months (95% CI: 29.1–35.6 months), 75 of 108 (69%), patients had progressed and 57 of 108 (53%) had died. Median PFS was 12.0 months (95% CI: 9.1–14.9 months) and median OS was 21.1 months (95% CI: 11.4–30.7 months). In univariate analyses, PFS and OS were independent of age, gender, tissue genotype at diagnosis, and previous EGFR TKI therapy but correlated with presence of extra-thoracic metastases (Table 2). Detectable plasma-based ctDNA was associated with shorter PFS (median 8.4 versus 29.4 months, HR 4.26, 95% CI: 2.55–7.10, P<0.0001) (Table 2 and Figure 2A) and OS (median 15.3 versus not reached, HR 3.23, 95% CI: 1.80–5.78, P<0.0001) (Table 2 and Figure 2B). Multivariate analysis revealed that ctDNA status was independently associated with PFS and OS (HR 4.87, 95% CI: 2.81–8.44, P<0.0001; HR 3.49, 95% CI: 1.88–6.50, P<0.0001) (Table 2).
Full table
Within eight weeks after osimertinib initiation, activating EGFR mutations and the T790M mutation were detected in plasma ctDNA of 19/57 (33%) and 8/57 (14%) patients, respectively. The C797S mutation was first detected 5.7 months after osimertinib initiation and was found in 6 of 57 (11%) patients.
We observed no association between clinical features and presence or absence of activating EGFR mutations (data not shown). Patients with persistence of the activating EGFR mutation had a significantly lower RR than patients without detectable activating EGFR mutations (24.5% versus 75.5%, P=0.001) (Table 3). Presence or absence of the T790M mutation had no impact on response to osimertinib (Table 3).
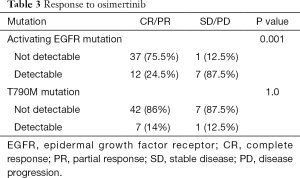
Full table
Patients with persisting activating EGFR mutations in ctDNA within eight weeks after the first dose of osimertinib had a significantly shorter PFS (median 3.4 versus 26.9 months; HR 6.17, 95% CI: 3.03–12.56, P<0.0001) (Figure 2C and Table S1) and shorter OS (median 9.4 versus not reached; HR 4.83, 95% CI: 2.25–10.36, P<0.0001) (Figure 2D and Table S1) compared to patients with total clearance of the activating EGFR mutation in plasma. Similarly, presence of the T790M mutation in ctDNA correlated with shorter PFS (median 7.0 versus 19.0 months; HR 2.32, 95% CI: 1.00–5.37, P=0.05) and OS (median 16.0 versus 33.4 months; HR 2.76, 95% CI: 1.10–6.93, P=0.03) (Table S1). Multivariable analyses using stepwise backward elimination models showed that the persistence of activating EGFR mutations in plasma ctDNA was the only parameter that independently predicted shorter PFS (HR 7.83, 95% CI: 3.53–17.34, P<0.0001) and OS (HR 4.90, 95% CI: 2.25–10.69, P<0.0001) of patients (Table S1).
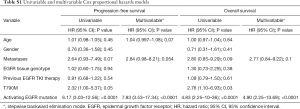
Full table
Discussion
The findings of our present study suggest that tracking of EGFR mutations in plasma ctDNA by means of ddPCR during second-line therapy with osimertinib is clinically relevant in patients with advanced EGFR-mutated NSCLC. If activating EGFR mutations persist in plasma ctDNA within eight weeks after start of osimertinib, a shorter PFS and OS can be expected in the respective patients.
Similar findings have been reported by others (9-11). In an exploratory analysis of the FLAURA trial (11), persistence of activating EGFR mutations in ctDNA at three weeks and six weeks after start of osimertinib therapy was associated with shorter PFS (11). In an exploratory analysis of the FASTACT-2 study, patients with EGFR mutation-negative plasma samples at cycle 3 had longer PFS and OS than patients whose samples were still EGFR mutation positive at cycle 3 (9). In another study, patients with a higher allele frequency of the activating EGFR mutation or a higher activating EGFR mutation/T790M ratio had a shorter PFS compared to those with a lower allele frequency or a lower ratio (10).
Our results suggest that EGFR mutation tracking could be useful for guiding treatment in the future. Patients in whom the activating EGFR mutations in ctDNA are not found within eight weeks after osimertinib initiation, should continue with osimertinib. In these patients, a median PFS of 26.9 months was observed and median OS survival was not reached at a follow-up of 34.8 months. However, patients with persisting activating EGFR mutations in plasma ctDNA may require a change in treatment because of their poor outcome. They may benefit from the addition of chemotherapy to osimertinib or other treatments.
This possibility is supported by findings of phase III trials in the first-line setting, in which the combination of gefitinib with chemotherapy resulted in longer PFS and OS compared to gefitinib alone (13,14). Therefore, these treatment strategies warrant further investigation within clinical trials among patients with persistence of activating EGFR mutations.
Another treatment strategy in patients with persisting activating EGFR mutations could be chemo-immunotherapy (15). This treatment option is supported by an exploratory analysis of the IMpower150 trial, which indicated longer OS for the addition of atezolizumab to chemotherapy plus bevacizumab compared to chemotherapy plus bevacizumab in patients with advanced non-squamous EGFR mutant NSCLC (15).
Our findings indicate that EGFR mutation tracking during second-line osimertinib therapy provides clinically relevant information. Patients with persistence of activating EGFR mutations in plasma ctDNA eight weeks after the first dose of osimertinib have worse survival and may require other treatments such as the combination of osimertinib with chemotherapy or chemo-immunotherapy. All these treatment options should be explored within clinical trials in the future and may further improve the outcome of patients with advanced EGFR-mutated NSCLC.
Acknowledgments
Funding: The study was funded by AstraZeneca Austria.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr.2020.03.02). AB reports personal fees from AstraZeneca, outside the submitted work; MJH reports personal fees from AstraZeneca, personal fees from Boehringer Ingelheim, personal fees from Bristol-Myers Squibb, personal fees from Merck Sharp & Dohme, personal fees from Novartis, personal fees from Pfizer, personal fees from Roche, outside the submitted work; RP reports personal fees from AstraZeneca, personal fees from Boehringer Ingelheim, personal fees from Gedeon Richter, personal fees from Genmab, personal fees from Merck Sharp & Dohme, personal fees from Regeneron, personal fees from Roche, outside the submitted work; RP serves as the unpaid editorial board member of Translational Lung Cancer Research from Dec 2019 to Nov 2020. MF reports personal fees from AstraZeneca, personal fees from Bayer, personal fees from Biomedica, personal fees from Boehringer Ingelheim, personal fees from Eli Lilly, personal fees from Merck Sharp & Dohme, personal fees from Myriad Genetics Inc., personal fees from Pfizer, personal fees from Roche, outside the submitted work; US has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The collection and analysis of blood samples was approved by the local ethics committee (EK No. 1132/2016) and informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Buder A, Tomuta C, Filipits M. The potential of liquid biopsies. Curr Opin Oncol 2016;28:130-4. [Crossref] [PubMed]
- Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res 2014;20:1698-705. [Crossref] [PubMed]
- Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol 2016;2:1014-22. [Crossref] [PubMed]
- Buder A, Hochmair MJ, Schwab S, et al. Cell-Free Plasma DNA-Guided Treatment With Osimertinib in Patients With Advanced EGFR-Mutated NSCLC. J Thorac Oncol 2018;13:821-30. [Crossref] [PubMed]
- Hochmair MJ, Buder A, Schwab S, et al. Liquid-Biopsy-Based Identification of EGFR T790M Mutation-Mediated Resistance to Afatinib Treatment in Patients with Advanced EGFR Mutation-Positive NSCLC, and Subsequent Response to Osimertinib. Target Oncol 2019;14:75-83. [Crossref] [PubMed]
- Buder A, Setinek U, Hochmair MJ, et al. EGFR Mutations in Cell-free Plasma DNA from Patients with Advanced Lung Adenocarcinoma: Improved Detection by Droplet Digital PCR. Target Oncol 2019;14:197-203. [Crossref] [PubMed]
- Mok T, Wu YL, Lee JS, et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin Cancer Res 2015;21:3196-203. [Crossref] [PubMed]
- Del Re M, Bordi P, Rofi E, et al. The amount of activating EGFR mutations in circulating cell-free DNA is a marker to monitor osimertinib response. Br J Cancer 2018;119:1252-8. [Crossref] [PubMed]
- Zhou C, Imamura F, Cheng Y, et al. Early clearance of plasma EGFR mutations as a predictor of response to osimertinib and comparator EGFR-TKIs in the FLAURA trial. J Clin Oncol 2019;37:abstr 9020.
- Gray JE, Peled N, Markovets A, et al. Longitudinal circulating tumour DNA (ctDNA) monitoring for early detection of disease progression and resistance in advanced NSCLC in FLAURA. Ann Oncol 2019;30:LBA17. [Crossref]
- Nakamura A, Inoue A, Morita S, et al. Phase III study comparing gefitinib monotherapy (G) to combination therapy with gefitinib, carboplatin, and pemetrexed (GCP) for untreated patients (pts) with advanced non-small cell lung cancer (NSCLC) with EGFR mutations (NEJ009). J Clin Oncol 2018;36:abstr 9005.
- Noronha V, Patil VM, Joshi A, et al. Gefitinib Versus Gefitinib Plus Pemetrexed and Carboplatin Chemotherapy in EGFR-Mutated Lung Cancer. J Clin Oncol 2020;38:124-36. [Crossref] [PubMed]
- Reck M, Mok TSK, Nishio M, et al. Atezolizumab plus bevacizumab and chemotherapy in non-small-cell lung cancer (IMpower150): key subgroup analyses of patients with EGFR mutations or baseline liver metastases in a randomised, open-label phase 3 trial. Lancet Respir Med 2019;7:387-401. [Crossref] [PubMed]