
Novel ALK-specific mRNA in situ hybridization assay for non-small-cell lung carcinoma
Introduction
Anaplastic lymphoma receptor tyrosine kinase (ALK) gene rearrangements occur in 4–5% of patients with lung adenocarcinoma, and these patients respond to ALK tyrosine kinase inhibitors (1-4). Although ALK break-apart fluorescence in situ hybridization (FISH) is the standard for detecting ALK rearrangements in non-small-cell lung carcinoma (NSCLC), the assay is technically challenging and costly. In contrast, immunohistochemistry (IHC) is a cost-effective, widely utilized screening method used routinely in most pathology laboratories. Given the lack of ALK protein expression in normal lung tissue, ALK-IHC would seem to be an ideal technique for detecting ALK-positive NSCLC. However, the ALK-IHC method is limited by variabilities in antibody sensitivity and inter-observer agreement (5,6), although recent work with commercially available ALK antibodies suggests that the IHC-based tests represent a reliable screening strategy (7,8).
Recently, multiplexed gene-sequencing panels have become preferred over multiple single-gene tests for identifying other treatment options beyond targeted therapies against mutant EGFR, ALK, and ROS1 (9,10). Next generation sequencing (NGS) has changed the practice of molecular diagnostics considerably for lung cancer. Substantial resources are necessary for clinical NGS implementation, and the assays are complex in terms of design, performance, and interpretation. Consequently, this technology is not universally employed (8).
Recent technical advances in mRNA in situ hybridization (mRNA-ISH) enable detection of cellular mRNA in formalin fixed paraffin embedded (FFPE) tissue using specific target probes with increased sensitivity and reduced background noise (11). In addition, mRNA-ISH can be fully automated as a bright-field mRNA-ISH assay using conventional bright field light microscopy. However, ALK mRNA-ISH is not an established clinical diagnostic tool, and its practical utility has not been evaluated. Here, we studied ALK mRNA expression in NSCLC by mRNA-ISH and compared its performance with that of IHC and FISH for examining the status of ALK rearrangements.
Methods
Patients and specimens
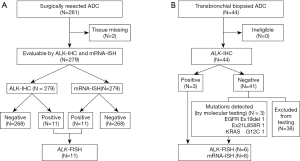
The study included 281 patients who underwent surgical lung adenocarcinoma resection at Asahikawa Medical University Hospital from February 2001 to March 2013, as well as 44 patients who underwent transbronchial-biopsied (TBB) after diagnosis with primary lung adenocarcinoma from January to October 2015 (Figure 1). Histological determination was based on the multidisciplinary classification system for lung adenocarcinoma proposed by the International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society (IASLC/ATS/ERS) in 2011 (12). Pathological stage was determined according to the UICC TNM Classification of Malignant Tumors (8th edition) (13,14). The clinical data were obtained from medical records retrospectively. Due to the retrospective nature of the study informed consent was not required. The study protocol was approved by the Asahikawa Medical University Research Ethics Committee (#19056).
Methodology
mRNA-ISH was performed for all surgically resected samples. FISH was used to assess cases that were either IHC- or mRNA-ISH-positive. In TBB samples, mRNA-ISH and FISH were applied with several representative cases, including ALK-IHC-positive samples and samples with other mutations. Using mRNA-ISH, we assessed all specimens that stained positive with ALK-specific antibodies. We also evaluated samples from patients with common EGFR mutations (exon 19, codon E746–A750 deletion mutation; exon 21, L858R point mutation) or KRAS mutations, which we verified by a highly sensitive allele-specific polymerase chain reaction (PCR)-based method using an ALK-negative control. Six samples with mutations (three in ALK, two in EGFR, and one in KRAS) were analyzed.
mRNA-ISH
Tissue microarrays (TMAs) were prepared from the most representative sections of each FFPE tumor block with a 5.0 mm diameter core. TMAs were prepared with a thickness of 4 m immediately prior to mRNA-ISH. A dedicated slide rack and oven were used to maintain a constant temperature and humidity during the hybridization and amplification procedures. In this study, we employed a commercially available mRNA-ISH kit (RNAscope 2.0 FFPE Assay-RED; Advanced Cell Diagnostics, CA, USA) and a pre-designed probe (catalog number 314191, Hs-ALK-E19-29, Advanced Cell Diagnostics) that specifically labelled ALK mRNA at the sequence encoding the ALK tyrosine kinase domain. Fast Red dye was used for chromogenic staining to avoid false positives resulting from high lung pigmentation or anthracosis. A standard bright-field microscope (Olympus U-TV0.5×C-3, Japan) was used for observations.
Target mRNA was visualized in the nucleus or cytoplasm as a red spot or cluster. According to the protocol for the RNAscope 2.0 FFPE Assay-RED provided by the manufacturer, the mRNA-expression level can be categorized into five grades: 0 (no staining or <1 dot/cell); 1 (1–3 dots/cell), 2 (4–10 dots/cell), 3 (>10 dots/cell or <10% positive cells with dot clusters), and 4 (>10 dots/cell or >10% positive cells with dot clusters). The manufacturer’s protocol suggests that users may need to scale the criteria according to gene-expression levels. Because accurate counting of the number of dots was difficult, we modified the protocol as follows: 0 (no staining, 0 dots), 1 (sparse dots, not homogeneously spread throughout the tumor cells, with no clusters), 2 (a moderate number of dots, which may or may not have been homogeneously spread throughout the tumor cells, with no clusters), 3 (moderate numbers of dots with a few clusters), and 4 (homogeneous distribution of clusters in the tumor cells). A cluster was defined as an agglomerate of dots in which individual dots could not be clearly defined. Scores of 1–4 were considered to be mRNA-ISH-positive, and a score of 0 was considered mRNA-ISH-negative. Samples with mRNA-ISH scores of 1–4 were analyzed by FISH. Cases in which the dots or clusters were located only at the edge of the tissue, stroma, intra-alveolar macrophages, or discharges were considered to be negative.
IHC analysis
TMAs and TBB samples, sectioned at a thickness of 4 m, were stained with a rabbit anti-ALK monoclonal primary antibody (Clone D5F3, Cell Signaling Technology, Danvers, MA, USA catalog number 3633, 1:500 dilution) in Dako REAL Antibody Diluent, using an automated IHC stainer (Leica Bond-III, Dako, Trappes, France). IHC results were scored based on the staining intensity and the proportion of stained cells. The staining-intensity categories were: 0 (no staining), 1 (mild staining), 2 (moderate staining), and 3 (heavy staining). The proportion of stained cells in each category was: 0 (no cells stained), 1 (1–20% of tumor cells stained), 2 (21–50% of tumor cells stained), and 3 (>51% of tumor cells stained). If both the intensity and proportion scores were 1 or more, the sample was defined as IHC-positive. Two common EGFR mutations (exon 19, codon E746–A750 deletion and exon 21, L858R point mutation) were examined in all TMA and TBB samples using rabbit EGFR mutation-specific monoclonal antibodies [clone D6B6 (catalog number 2085) and clone 43B2 (catalog number 3197), respectively; Cell Signaling Technology].
ALK-FISH
mRNA-ISH-positive samples or IHC-positive samples were consecutively analyzed by ALK break-apart FISH (Vysis ALK Break Apart FISH Probe Kit, Abbott Molecular, Il, USA) according to the manufacturer’s protocol. Serial 4 µm-thick FFPE sections were used for analysis. Over 50 cancer cells were scored for each sample, and the signals were evaluated as previously described (15) according to the IASLC guidelines for ALK testing (16). FISH was defined as positive when the split pattern and a single red signal (single 3' pattern) occurred in >15% of the tumor cells examined.
Patient and sample exclusion
Samples were excluded from analysis in cases where missing tissue, no staining, or poor tissue quality were observed. Two patients’ tissues were excluded because they expired during storage. On this basis, 279 surgical specimens were evaluable, and 44 TBB specimens included adequate tumor tissue (diagnosed as primary lung adenocarcinoma) that was assessable by ALK-IHC.
Treatment with ALK tyrosine kinase inhibitors
Patients with ALK-rearranged, advanced-stage NSCLC were treated with ALK tyrosine kinase inhibitors in accordance with standard therapy in Asahikawa Medical University Hospital (17-19). Three patients were treated with crizotinib (Xalkori, Pfizer, NY, USA), one of which received alectinib (Alecensa, Chugai Pharma, Tokyo, Japan) treatment after acquiring resistance to crizotinib.
Statistical analysis
The concordance between mRNA-ISH and IHC data was calculated using kappa statistics. The kappa coefficients were interpreted as follows: >0.81, high agreement; 0.61–0.80, good to fair agreement; 0.41–0.60, moderate agreement; 0–0.40, poor agreement. The correlation between the mRNA-ISH score and IHC score was analyzed by determining Pearson’s correlation coefficient.
Results
Surgically resected samples
Patient and tumor characteristics
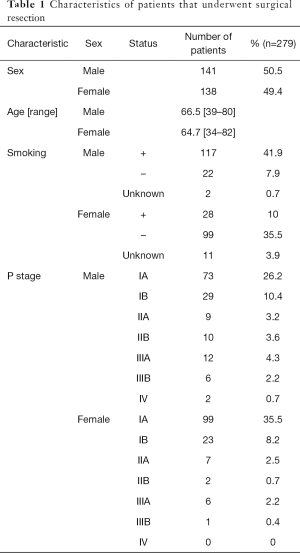
The characteristics of the 279 surgically resected tumors are presented in Table 1. In summary, the majority of male patients had a smoking history, and the majority of female patients were never smokers. Most patients (80.3%) had pathological stage I NSCLC at the time of analysis, and two patients (0.7%) were classified as having stage IV NSCLC based on the presence of pleural metastases.
Full table
IHC analysis
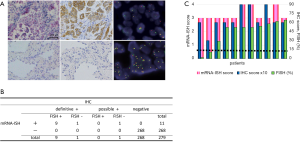
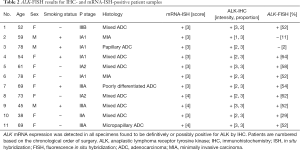
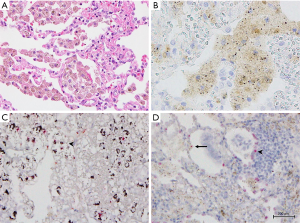
ALK protein expression was evaluable in 279 samples. ALK expression was observed in 11 tumor samples (3.9%). Representative micrographs of ALK-positive and ALK-negative cases are shown in Figure 2. The mRNA-ISH, IHC, and FISH results for these 11 cases are summarized in Table 2. Ten tumor specimens had obvious cytoplasmic staining, with intensity and proportion scores of 2 or 3. We classified these cases as being definitively ALK-IHC. One sample (patient No. 2) displayed mild staining of all tumor cells, with intensity and proportion scores of 1 and 3, respectively. In patient No. 2, stippling in alveolar macrophages or extracellular mucin coexisted equally with highly granular staining (Figure 3A,B). Although this specimen may have been mis-characterized with a false-positive IHC result, we considered it to be probably ALK-IHC-positive based on the mild staining occurring in most tumor cells. No cases with a proportion score of 1 were observed. The two common EGFR mutations were not identified by IHC in these 11 cases. The remaining 268 samples were not stained and were classified as being ALK-IHC-negative.
Full table
ALK mRNA-ISH data
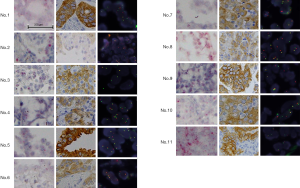
Representative micrographs of ALK mRNA-ISH-positive and ALK mRNA-ISH-negative samples are shown in Figure 2A. Intracellular red dots or clusters were observed in cases where ALK mRNA was present. We found that 279 specimens were evaluable by ALK mRNA-ISH, and we identified intracellular red signals in 11 specimens (3.9%), which were also classified as definitively and possibly IHC-positive cases (Figure 2B). Figure 2C shows correlations between the mRNA-ISH and IHC scores, and the percentages of FISH-positive cells in these 11 specimens. For the mRNA-ISH-positive cases, the correlation coefficient for the relationship between mRNA-ISH and IHC scores was moderate (0.51). In one case (patient No. 2), an average of less than 1 dot/tumor cell was observed. In this case, some clusters were also observed, and this specimen was assigned a score of 3 according to our criteria (Figure 3C,D). In the remaining 10 cases, dots or clusters were distributed homogenously in the tumor cells and were clearly recognizable, corresponding to scores of 3 or 4 (Figure 4). These 11 cases were designated as mRNA-ISH-positive and were further evaluated by FISH. Dots or clusters were not detected in the remaining 268 samples, which were all designated as mRNA-ISH-negative.
ALK-FISH data
ALK break-apart FISH was conducted on 10 definitively IHC-positive samples and 1 possibly IHC-positive sample. Nine specimens were judged to be ALK-FISH-positive, which was in complete agreement with the IHC designations. The remaining two (patients No. 2 and 3) were found to have 11% and 2% FISH-positive cells, respectively, and were judged to be ALK-FISH negative. The majority of the definitive and possible ALK-IHC-positive cases (81.8%) matched the FISH results. When the possibly IHC-positive case (patient No. 2) was excluded, there was a 90% match with the FISH results.
Correlations between the mRNA-ISH, IHC, and FISH data
Using the IHC data as a reference, the RNAscope®ALK mRNA-ISH assay had a sensitivity and specificity of 100% (κ=1.0), according to our modified mRNA-ISH criteria. According to the manufacturer’s instructions, patient No. 2 should be considered mRNA-ISH-negative and possibly IHC-positive; using these criteria, the sensitivity and specificity were 90.9% and 100%, respectively (κ=0.9505). The results for the mRNA-ISH-positive patients, including patient No. 2, showed 81.8% agreement with the FISH results, with mRNA-ISH detecting ALK mRNA in two ALK-FISH-negative specimens. The sample from patient No. 3 showed homogeneously distributed dots and clusters by mRNA-ISH testing, and non-uniform IHC staining (Figure 5), whereas the ALK-FISH test was negative. The tumor from patient No. 2 was ALK-FISH-negative, but showed weak homogenous staining by IHC, and displayed dots and clusters in tumor cells by mRNA-ISH. The remaining 9 cases demonstrated unequivocal IHC staining and obvious mRNA-ISH signals, and positive ALK-FISH results.
TBB samples
IHC analysis and patient characteristics
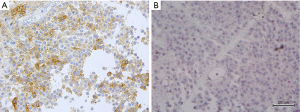
ALK protein expression was evaluable in 44 TBB adenocarcinomas. Cytoplasmic staining was observed in tumor samples from three patients (6.8%; patients No. 1–3), with moderately to markedly intense staining in a large proportion of cells (Table 3, Figure 6). We determined that these three samples were definitively IHC-positive. All IHC-positive samples were not stained with either of two EGFR mutation-specific antibodies. The remaining 41 samples were IHC-negative.

Full table
The characteristics of six TBB specimens analyzed by mRNA-ISH, IHC, and FISH are shown in Table 3. Two ALK-IHC-positive patients had early-stage NSCLC and underwent surgical lobectomy. One ALK-IHC-positive patient had advanced-stage NSCLC at the time of diagnosis and was treated with crizotinib. Histopathological examination revealed that two of three ALK-IHC-positive adenocarcinomas contained micropapillary components. Two patients with EGFR mutations (patients No. 4 and 5) were female with no smoking history. One patient with a KRAS mutation (patient No. 6) was a young female with a heavy smoking history.
ALK-FISH data
ALK break-apart FISH was conducted on six TBB adenocarcinomas, including three ALK-IHC-positive and three ALK-IHC-negative samples (two EGFR mutations and one KRAS mutation were confirmed by high-sensitivity quantitative PCR). Three samples were ALK-FISH-positive, in agreement with the IHC results. In the two samples with a KRAS mutation or a deletion in exon 19 of EGFR (codons E756–A750), 8% of the cells were ALK-FISH-positive. These two cases were judged to be ALK-FISH-negative. The ALK-expression status in the sample with an EGFR exon 21 L858R point mutation was judged to be indeterminant.
ALK mRNA-ISH data
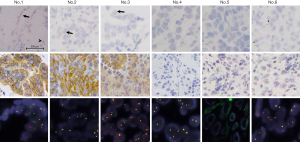
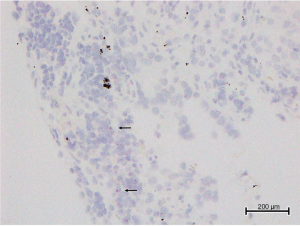
ALK mRNA-ISH was evaluable in six TBB samples. Representative micrograph images of the six TBB specimens analyzed by ALK mRNA-ISH, IHC, and FISH are shown in Figure 6. ALK mRNA expression was clear in one case (patient No. 1, Figure 7), which was strongly positive by IHC and ALK-FISH. In this specimen, dots were located homogeneously in the lepidic tumor cells, with some clusters (Figure 6). We designated this specimen as being mRNA-ISH-positive, with a score of 3. In the two remaining IHC-positive samples (patients No. 2 and 3), sparse dots were observed at an average of less than 1 dot/tumor cell. Dots were distributed evenly throughout the tumor tissue, and no clusters were evident. These two specimens were assigned an mRNA-ISH score of 1 and were considered mRNA-ISH positive according to our modified criteria.

In two samples with EGFR mutations (patients No. 4 and 5), dots or clusters were not observed in the tumor cells, and these samples were considered mRNA-ISH negative. In the sample with KRAS mutation (patient No. 6), some sparsely distributed dots were detected in the tumor cells, predominantly at the edge of the tissue (Figure 8), and no clusters were observed. This sample was considered mRNA-ISH-positive with a score of 1 according to our modified criteria.
Correlations between the IHC, FISH, and mRNA-ISH data
The sensitivity and specificity of mRNA-ISH could not be assessed with the TBB samples. Analysis of six samples by mRNA-ISH, FISH, and IHC revealed that ALK signals were definitively detected by mRNA-ISH in all FISH- and IHC-positive samples. However, cluster formation was less prominent in the TBB samples than in the surgical specimens.
Discussion
In our study, ALK-IHC and ALK mRNA-ISH were evaluated with 279 surgically resected lung adenocarcinomas, and break-apart FISH was examined for IHC- or mRNA-ISH-positive cases. The ALK-IHC-positive and mRNA-ISH-positive samples were identical and accounted for 3.9% of all samples tested. ALK-FISH-positive results were confirmed in 9 out of 11 IHC- and mRNA-ISH-positive cases. A relatively high frequency of ALK rearrangements (4–9%) has been reported in NSCLC (1-4). In our study, the frequency of ALK protein expression was similar to the results obtained previously with the ALK-specific D5F3 antibody, which was reported to have a high sensitivity and specificity (5,7). Pekar-Zlotin et al. reported that false-negative samples were observed with IHC (with NGS as the gold standard), and the sensitivity and specificity of IHC were found to be 100% and 97.7% respectively (20). Clinical NGS implementation has been widely employed for multiplex genetic testing; however, the quality and quantity of the extracted DNA or RNA are critical. It can be difficult to obtain adequate DNA or RNA from biopsy samples. Because our mRNA-ISH method yielded a high correlation rate with ALK-IHC-positive samples, mRNA-ISH testing may be a viable method for testing samples that are IHC-negative from patients with clinical characteristic that are highly associated with ALK-positive results, such as onset at an early age or a history of being a never smoker.
mRNA-ISH showed high sensitivity and specificity with surgically resected lung specimens, when using IHC as a reference. Moreover, all FISH-positive surgical specimens and TBB specimens were also mRNA-ISH positive. These results highlight the utility of mRNA-ISH for detecting ALK expression in NSCLC. With mRNA-ISH, the ALK signal distribution and intensity were uniform in all but two surgically resected samples with false-positive FISH/IHC signals that exhibited a non-homogeneous signal distribution, similar to the IHC results. Although ALK rearrangements and KRAS mutations are regarded as mutually exclusive (21), we detected some tumor cells that expressed ALK in one KRAS-mutated TBB sample. Additional testing by reverse transcription-PCR or deep sequencing may be required to investigate this sample. The mRNA-signal intensities in TBB specimens were relatively weak compared with those in surgical specimens, even though the IHC staining intensities were similar. These differences may reflect RNA-quality issues related to the tumor size and time because the tissues were sectioned. To improve the mRNA-signal intensity, we recommend performing the assays immediately after the initial tissue slices are prepared for histological diagnosis.
In our study, we detected cellular ALK mRNA expression in FFPE tissues using specific target probes against ALK mRNA encoding exon 19–exon 29 lesions. Nonetheless, compared to FISH and IHC, it was reasonable to question whether mRNA-ISH was sufficiently sensitive to detect ALK rearrangements. Moreover, it was unclear whether ALK mRNA and ALK protein expression are correlated quantitatively. Although the processes regulating the transcription and translation of ALK-fusion genes are not fully understood, ALK fusions may strongly correlate with increased ALK mRNA-expression levels (22). To date, limited mRNA-ISH data related to lung cancer have been published, and the appropriate scoring and interpretation systems remain controversial. The IHC and mRNA-ISH scores demonstrated a very weak quantitative correlation in our study, and establishing appropriate cut-off settings will be important challenges for future studies.
In summary, the ALK mRNA-ISH data were highly correlated with the IHC data, and ALK mRNA-ISH identified ALK mRNA expression in every FISH-positive sample. ALK mRNA-ISH can be conducted using FFPE samples, can detect unknown fusions, offers a short turnaround time, can be adopted at numerous facilities, and can be used to confirm the presence of tumor cells. The limitations of this study are that the results may vary significantly depending on the FFPE quality, and RNA of fair quality is required. We conclude that mRNA-ISH can potentially complement NSCLC diagnosis based on detecting ALK rearrangements in cases where IHC staining is negative, but the clinical characteristics are highly associated with ALK-positive results, such as early-onset disease or a history of never smoking history.
Acknowledgments
We would like to thank Editage for English language editing.
Funding: This study was supported by a Grant-in-Aid for Young Scientists (B) (grant number 26860595) from the Ministry of Education, Culture, Sports, Science and Technology.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr.2020.03.04). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Due to the retrospective nature of the study informed consent was not required. The study protocol was approved by the Asahikawa Medical University Research Ethics Committee (#19056).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sasaki T, Rodig SJ, Chirieac LR, et al. The biology and treatment of EML4-ALK non-small cell lung cancer. Eur J Cancer 2010;46:1773-80. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Camidge DR, Bang YJ, Kwak EL, et al. Activity and safety of crizotinib in patients with ALK-positive non-small-cell lung cancer: updated results from a phase 1 study. Lancet Oncol 2012;13:1011-9. [Crossref] [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. [Crossref] [PubMed]
- Mino-Kenudson M, Chirieac LR, Law K, et al. A novel, highly sensitive antibody allows for the routine detection of ALK-rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res 2010;16:1561-71. [Crossref] [PubMed]
- Takeuchi K. Interpretation of anti-ALK immunohistochemistry results. J Thorac Oncol 2013;8:e67-8. [Crossref] [PubMed]
- Conklin CM, Craddock KJ, Have C, et al. Immunohistochemistry is a reliable screening tool for identification of ALK rearrangement in non-small-cell lung carcinoma and is antibody dependent. J Thorac Oncol 2013;8:45-51. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol 2018;13:323-58. [Crossref] [PubMed]
- Drilon A, Wang L, Arcila ME, et al. Broad, hybrid capture–based next-generation sequencing identifies actionable genomic alterations in lung adenocarcinomas otherwise negative for such alterations by other genomic testing approaches. Clinical cancer research 2015;21:3631-9. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Beasley MB, et al. Molecular Testing Guideline for Selection of Lung Cancer Patients for EGFR and ALK Tyrosine Kinase Inhibitors: Guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Mol Diagn 2013;15:415-53. [Crossref] [PubMed]
- Wang F, Flanagan J, Su N, et al. RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 2012;14:22-9. [Crossref] [PubMed]
- Travis WD, Brambilla E, Noguchi M, et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011;6:244-85. [Crossref] [PubMed]
- Brierley JD, Gospodarowicz MK, Wittekind C. TNM classification of malignant tumours. John Wiley & Sons, 2016.
- Nicholson AG, Chansky K, Crowley J, et al. The International Association for the Study of Lung Cancer Lung Cancer Staging Project: Proposals for the Revision of the Clinical and Pathologic Staging of Small Cell Lung Cancer in the Forthcoming Eighth Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:300-11.
- Rodig SJ, Mino-Kenudson M, Dacic S, et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res 2009;15:5216-23. [Crossref] [PubMed]
- Tsao M, Hirsch F, Yatabe Y. IASLC atlas of ALK testing in lung cancer. Aurora, CO: IASLC Press, 2013.
- Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167-77. [Crossref] [PubMed]
- Peters S, Camidge DR, Shaw AT, et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:829-38. [Crossref] [PubMed]
- Hida T, Nokihara H, Kondo M, et al. Alectinib versus crizotinib in patients with ALK-positive non-small-cell lung cancer (J-ALEX): an open-label, randomised phase 3 trial. Lancet 2017;390:29-39. [Crossref] [PubMed]
- Pekar-Zlotin M, Hirsch FR, Soussan-Gutman L, et al. Fluorescence in situ hybridization, immunohistochemistry, and next-generation sequencing for detection of EML4-ALK rearrangement in lung cancer. Oncologist 2015;20:316-22. [Crossref] [PubMed]
- Takahashi T, Sonobe M, Kobayashi M, et al. Clinicopathologic features of non-small-cell lung cancer with EML4-ALK fusion gene. Ann Surg Oncol 2010;17:889-97. [Crossref] [PubMed]
- Zhang X, Zhang S, Yang X, et al. Fusion of EML4 and ALK is associated with development of lung adenocarcinomas lacking EGFR and KRAS mutations and is correlated with ALK expression. Mol Cancer 2010;9:188. [Crossref] [PubMed]


