
From precision medicine to imprecision medicine through limited diagnostic ability to detect low allelic frequency mutations
The Rosetta Stone of the new era of biomedicine includes five essential ideas to understand the real concept of precision medicine (PM): preventive, predictive, personalized, participatory (1), and pertinent. The application of these five “Ps” is currently the Holy Grail of the clinical routine and particularly important for cancer are the concepts of prediction and personalization. It is especially significant in those diseases characterized by higher biological heterogeneity, as it is the case of lung cancer. Obviously, an improvement of biological knowledge must be accompanied by technological breakthroughs. In the last years, new molecular analysis technologies characterized by high specificity and sensitivity rates are being developed, allowing detection of rare clonal sub-types with low allelic frequency. However, the specificity and sensitivity present high variability among platforms, what is of particular importance for the detection of very low allelic frequency mutations. In fact, it has been accepted that variants with a low allele frequency affect the accuracy of mutation detection methodologies (2). Moreover, the variability in the limits of detection (LoD) is especially important for a correct treatment choice as well as an efficient stratification of patients susceptible to be treated with a particular therapy. Therefore, selection of methodologies with ability to discriminate between high and low allelic frequencies is essential for a correct application of PM. In the case of lung cancer, there are several mutation-targeting strategies for personalized treatments among which, tyrosine kinase inhibitors (TKI) are a widely used anti-cancer strategy. The key target of these drugs is the EGFR gene that, when overexpressed, activates cell survival and proliferation pathways. The success or failure of TKI treatments usually relies on presence or absence of sensitizing mutations respectively but together with them also treatment-resistant mutations might play a role (3). The most common resistance mutation is EGFR T790M so screening for this mutation offers information on whether the patient is candidate or not for TKI treatments.
In this issue, Lettig et al. (4) compared the specificity and sensitivity of three different technologies to assess somatic EGFR mutations: matrix-assisted laser time-of-flight desorption/ionization (MALDI-TOF) mass spectrometry (MS), allele-specific real-time PCR (AS-PCR) and droplet digital PCR (ddPCR). Likewise, they developed ultra-deep next generation sequencing (NGS) validation of the identified mutations in EGFR in non-small cell lung cancers (NSCLCs) using SiRe® panel. The findings of this work are not only technologically but also clinically relevant. From the technological point of view, the work confirms results obtained by other groups, demonstrating that MALDI-TOF does not offer the higher sensitivity required to detect somatic mutations at allelic frequencies below 5% (5). In this context, the approach of numerous studies is to evaluate the mutation-detection ability of different platforms, with varying sensitivities, what conditions the correct treatment choice, and might transform “PM” into an “imprecision medicine” (6). In this paper, Lettig et al. demonstrated that ddPCR offers good sensitivity (7), tissue economy and quick turnaround time compared to MALDI-TOF, three aspects that are required to guide treatment decision (8). In addition, the work of Lettig et al. reflects the importance of correctly select the most sensitive methodology, since they identified presence of different clones harbouring concurrent mutations with varying mutated allelic frequencies (mAF) affecting EGFR. Despite not being a novel concept, the presence of concurrent mutations in EGFR has been described as an important factor for development of treatment resistance (9,10). In fact, according to the authors, presence of these subclones conditions overall survival (OS) and progression-free survival (PFS). Nevertheless, their work has some limitations in terms of number of patients, which in turn, might bias the conclusions arising from their Kaplan-Meier curves. From this point of view, we argue that patients reported positive for T790M mutation might be split into two groups according to their mAF (either greater or lower than 5%) to more robustly determine the effect of each mutational load on treatment resistance. Therefore, treatment response will also depend on the heterogeneity present at the allele frequency level. Lettig et al. concluded that the presence of T970M with mAF higher than 5% prior TKI treatment identifies patients susceptible to be treated with third-generation EGFR-TKI in first line, however only three patients in their cohort fulfilled this criterion. On the contrary, they suggested that patients with low T970M mAF, together with concurrent sensitizing mutations (the majority of their cohort), might benefit from current treatments and would not need 3rd generation TKI in first line. Despite MALDI-TOF identified patients susceptible to be treated with 3rd generation TKI, it also raised the question whether patients assessed negative by MALDI-TOF should need to be analysed by other methodologies to rule out presence of mutations with lower than 5% allelic fraction. As the authors precisely suggest at the end of their discussion, the problem of acquired resistance might be due to “clonal selection” because of treatment selective pressure. Thus, it is imperative to standardize protocols and methodologies with enough sensitivity to detect all resistance mutations (including those at very low mAF) as these clones might be selected by the treatment, serving thus as prognostic and treatment response biomarkers. In fact, if these low mAF mutations are present since the beginning but the use of conventional methodologies is not able to detect them, then it may be possible that we treat patients with inadequate TKIs, thus offering “imprecise” rather than “precise” medicine (11).
Ultimately, the key point would be to determine which methodology, if only one is to be considered, might finally be the gold standard in the clinical routine, assuming that if two mutations are located within the same gene, any PCR-based method might have an amplification bias (competence between different substrates for PCR primers). Therefore, this limitation might affect mutation frequency calculation and in the case of ddPCR (despite its superior sensitivity), presence of false positives is regrettably an unwanted reality that could explain why for some patients, the mutation frequency was found very different between ddPCR and NGS. Consequently, it is undeniable that there is an urgent need to decide on the best methodologies to evaluate the genetic status associated to the set of targeted treatments currently available (Figure 1). Likewise, we cannot forget that these methodologies need to be accessible to a large number of hospitals, therefore are bound to be economically feasible (cancer affects wealthy and poor in the same way) and sensitive enough to analyze small amounts of DNA [e.g., circulating tumor DNA (ctDNA)], since response assessment involves the need to include liquid biopsies detection as a way of monitoring in real time the evolution of the disease and consequently, treatment response. Notably, the incorporation of liquid biopsies should be mandatory not just for monitoring but also in those cases in which tissue is not available (very common for NSCLC) or when tissue preservation might affect genetic results [e.g., formalin-fixed, paraffin-embedded (FFPE)]. In this line, some work is being done on the comparison of ctDNA and tissue mutation status using MALDI-TOF although further research is needed to extract adequate interpretations (12).
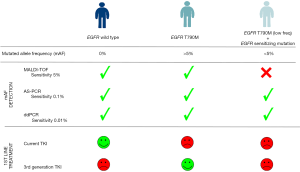
In conclusion, routine molecular testing in NSCLC is currently moving beyond the classical EGFR mutational analysis and the evolving technologies such as NGS or ddPCR, with superior sensibilities but also multiplexing capabilities, allow the evaluation of greater number of mutations/genes and thus, impact on disease management. The continuous progress of targeted therapies requires molecular testing for a wider panel of mutations and different technologies sensitive enough for mAF to be able to integrate personalized molecular diagnosis in the different Pathology departments. As a consequence, efficient testing of multiple molecular abnormalities is an urgent requirement in thoracic oncology not only for an efficient treatment administration but also for an improvement of the cost-benefit balance. Therefore, it is mandatory that numerous research groups tackle this challenge combining efforts to increase population sizes, homogenize protocols and standardize technologies so the 5 pillars of PM are translated into a reality in the clinical practice.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr.2020.03.07). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Auffray C, Charron D, Hood L. Predictive, preventive, personalized and participatory medicine: back to the future. Genome Med 2010;2:57. [Crossref] [PubMed]
- Wang JF, Pu X, Zhang X, et al. Variants with a low allele frequency detected in genomic DNA affect the accuracy of mutation detection in cell-free DNA by next-generation sequencing. Cancer 2018;124:1061-9. [Crossref] [PubMed]
- Jiang W, Cai G, Hu PC, et al. Personalized medicine in non-small cell lung cancer: a review from a pharmacogenomics perspective. Acta Pharm Sin B 2018;8:530-8. [Crossref] [PubMed]
- Lettig L, Sahnane N, Pepe F, et al. EGFR T790M detection rate in lung adenocarcinomas at baseline using droplet digital PCR and validation by ultra-deep next generation sequencing. Transl Lung Cancer Res 2019;8:584-92. [Crossref] [PubMed]
- Sherwood JL, Müller S, Orr MCM, et al. Panel Based MALDI-TOF Tumour Profiling Is a Sensitive Method for Detecting Mutations in Clinical Non Small Cell Lung Cancer Tumour. Lo AW, editor. PLoS One 2014;9:e100566.
- Ilie M, Hofman P. Pitfalls in Lung Cancer Molecular Pathology: How to Limit them in Routine Practice? Curr Med Chem 2012;19:2638-51. [Crossref] [PubMed]
- Jiang XW, Liu W, Zhu XY, et al. Evaluation of EGFR mutations in NSCLC with highly sensitive droplet digital PCR assays. Mol Med Rep 2019;20:593-603. [PubMed]
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the. Arch Pathol Lab Med 2018;142:321-46. [Crossref] [PubMed]
- Chen M, Xu Y, Zhao J, et al. Concurrent Driver Gene Mutations as Negative Predictive Factors in Epidermal Growth Factor Receptor-Positive Non-Small Cell Lung Cancer. EBioMedicine 2019;42:304-10. [Crossref] [PubMed]
- Santoni-Rugiu E, Melchior LC, Urbanska EM, et al. Intrinsic resistance to EGFR-Tyrosine Kinase Inhibitors in EGFR-Mutant Non-Small Cell Lung Cancer: Differences and Similarities with Acquired Resistance. Cancers (Basel) 2019;11:923. [Crossref] [PubMed]
- Shin HT, Choi YL, Yun JW, et al. Prevalence and detection of low-allele-fraction variants in clinical cancer samples. Nat Commun 2017;8:1377. [Crossref] [PubMed]
- Bonaparte E, Pesenti C, Fontana L, et al. Molecular profiling of lung cancer specimens and liquid biopsies using MALDI-TOF mass spectrometry. Diagn Pathol 2018;13:4. [Crossref] [PubMed]


