
ctDNA analysis reveals different molecular patterns upon disease progression in patients treated with osimertinib
Introduction
Epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) have been the standard of care for patients with advanced EGFR-mutant non-small-cell lung cancer (NSCLC) (1,2). However, most patients progress within 1 to 2 years (3). The EGFR p.T790M mutation is the most common resistance mechanism to first- and second-generation EGFR TKIs (4). Osimertinib, a third-generation TKI, has demonstrated its clinical efficacy in NSCLC tumors harboring the p.T790M mutation at disease progression after treatment with first- or second-generation EGFR TKIs (5). Moreover, in the randomized phase III FLAURA trial, osimertinib exceeded the standard of care gefitinib or erlotinib in treatment-naive NSCLC patients harboring EGFR exon 19 deletions and the p.L858R point mutation, giving rise to a significant improvement in median progression-free survival (PFS) compared with standard TKIs (6).
Nevertheless, acquired EGFR mutations conferring osimertinib resistance invariably emerge, such as the p.C797S mutation, which accounts for approximately 20–40% of the cases (7,8). Other resistance mechanisms have also been described (9,10). A better understanding of the diversity of mechanisms by which tumors acquire resistance to third-generation EGFR inhibitors is of particular relevance to the better clinical management of patients, making the analysis of circulating tumor DNA (ctDNA) during disease progression an attractive means of deriving new insights into tumor biology at different stages of the disease. In this paper, we describe an observational prospective cohort of 22 unselected patients treated with osimertinib with a median follow-up of 62 months. In addition, ctDNA analysis was performed on 326 samples collected throughout the course of disease.
Methods
Study cohort
The present observational study was conducted on 22 prospectively enrolled patients. Patients were followed from their diagnosis of stage IV disease. The study was approved by the Hospital Puerta de Hierro Ethics Committee and was conducted in accordance with the precepts of the Code of Ethics of The World Medical Association (Declaration of Helsinki). Written informed consent was obtained from all patient. Briefly, eligible patients were males and females with a pathologically confirmed diagnosis of stage IIIB–IV NSCLC tumor harboring an EGFR mutation, who were treated with a TKI, and who were candidates for receiving osimertinib. A complete staging workup was performed prior to recruitment. Data on demographic characteristics, clinicopathological features, tumor mutational status, vital status, disease status, drug dose adjustments and discontinuation of medication were collected in the study’s electronic database. Computed tomography (CT) measurements and magnetic resonance imaging (MRI) were obtained as clinically indicated. The clinical response was evaluated according to RECIST v1.1 criteria combined with a blinded medical judgment about the benefits of the treatment. Additionally, whole-body 18F-fluoro-2-deoxy-D-glucose positron emission tomography (18FDG-PET) CT scans were performed as clinically indicated.
Laboratory procedures
Three hundred and twenty-six whole blood samples were collected in an 8.5 mL PPT™ tube (Becton Dickinson Franklin Lakes, NJ, USA) containing a gel barrier to separate the plasma after centrifugation. Samples were processed as previously described (11-13). Briefly, after two consecutive centrifugations, cfDNA was isolated from plasma using the Maxwell® RSC (MR) ccfDNA Plasma Kit (Promega Corporation, Madison, WI, USA). The original EGFR-sensitizing mutation, and the p.T790M and p.C797S resistant mutations were analyzed by digital PCR (dPCR). Specifically, cfDNA was analyzed using commercially available predesigned TaqMan® Liquid Biopsy dPCR assays as well as custom TaqMan® assays in a QuantStudio® 3D Digital PCR System (Applied Biosystems, South San Francisco, CA, USA). dPCR reactions were carried out in a final volume of 18 µL and using 8.55 µL of cfDNA template. Subsequently, 14.5 µL were loaded into a QuantStudio 3D Digital PCR 20K chip. The cycling conditions were as follows: initial denaturation at 96 °C for 10 min, followed by 40 cycles at 56 °C for 2 min, and 98 °C for 30 s, a step of 72 °C for 10 min, and finally samples were maintained at 22 °C for at least 30 min. Chip fluorescence was measured twice. Results were analyzed with QuantStudio® 3D AnalysisSuiteTM Cloud Software. The automatic call assignments for each data cluster were manually adjusted when needed. The result of the assay is reported as the ratio of mutant DNA molecules relative to the sum of mutant and wild-type (wt) DNA molecules. A negative and a positive control DNA were included in every run.
Libraries were prepared using the OncomineTM Pan-Cancer Cell-Free Assay (Thermo Fisher, Palo Alto, CA, USA) according to manufacturer’s instructions. All the purifications were done using AMPure XP magnetic beads (Beckman Coulter, Inc., Brea, CA, USA). Library quantification was performed using the Ion Library TaqMan® Quantitation kit (Thermo Fisher, Palo Alto, CA, USA) in a StepOnePlusTM qPCR machine (Thermo Fisher, Palo Alto, CA, USA). The individual libraries were diluted to a final concentration of 100 pM. The final barcoded libraries were pooled and adjusted to a final concentration of 50 pM. Template preparation and chip loading were carried out on an Ion ChefTM System (Thermo Fisher, Palo Alto, CA, USA). Eight samples were loaded onto an Ion 550TM chip. Finally, Ion 550TM chips were sequenced in an Ion S5TM Sequencer (Thermo Fisher, Palo Alto, CA, USA).
Raw sequencing data were analyzed using Torrent Suite Software (v5.10.0). Sequencing coverage was analyzed using the Coverage Analysis (v.5.10.0.3) plug-in (Thermo Fisher, Palo Alto, CA, USA). Raw reads were aligned to the human reference genome hg19.
Variant calling, annotation and filtering were carried out on the Ion Reporter (v5.10) platform using the Oncomine TaqSeq Pan-Cancer Liquid Biopsy workflow (v5.10). The clinical significance of somatic variants was determined according to the Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer (14). Mutations with an allele frequency (AF) greater than or equal to 0.1% were considered positive.
Statistical analysis
Discrete variables are presented as frequencies and proportions, and continuous variables as means and standard deviations (SDs), unless otherwise specified. The median follow-up was estimated by the reverse Kaplan-Meier method (15). Overall survival (OS) and PFS were evaluated using the Kaplan-Meier survival function and Cox proportional hazards models. For OS analysis, time from the start of treatment with osimertinib to death or last follow-up was measured. PFS was defined as the time between the start of osimertinib treatment and disease progression, as assessed by RECIST criteria, or all-cause death. Patients who were alive on the last date of assessment and who had not experienced any event were censored at that time. Time to treatment discontinuation (TTD) of targeted therapy was defined as the time between the date when first-line treatment with a TKI began to the date of osimertinib discontinuation or death. Similarly, time to osimertinib discontinuation was also analyzed. Hazard ratios (HRs) were calculated from univariate Cox models. Significance was concluded for P values less than 0.05. Statistical analyses were performed using Stata 15.1 and R 3.1.2 software.
Results
Clinical outcomes
The study cohort included 22 patients. Clinico-pathological characteristics of the study population are presented in Table 1. The median age at diagnosis was 65 (range, 41–75) years. We found an unusually high prevalence of tobacco consumption, whereby 41% (9/22) of the patients were smokers (3/22) or former-smokers (6/22), with a mean consumption of 35 (SD: 28.5) pack-years. According to the pathologist’s report, 54.5% of the cases (12/22) harbored exon 19 deletions. In one case, a deletion in exon 19 co-occurred with the p.S768I mutation in exon 20. In addition, 45.5% (10/22) harbored the point mutation p.L858R in exon 21. These frequencies are as expected, based on previously published data. No significant differences were observed in OS and PFS with respect to the original EGFR-sensitizing mutation. At the start of osimertinib treatment, patients had a median of three metastatic sites, the most frequent locations being the lung (73%), the bone (64%), the pleura (59%), the central nervous system (23%) and the peritoneum (14%). The ECOG Performance Status varied from 0 to 2. Patients with an ECOG Performance Status of 0 exhibited improved OS (P=0.026).
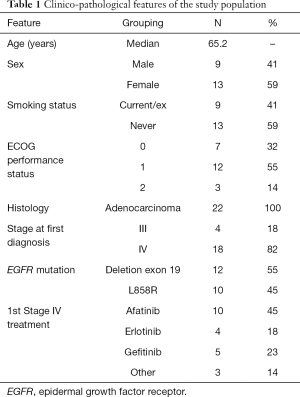
Full table
The median follow-up was 62 months. During the study, 12 deaths were recorded and progressive disease (PD) to osimertinib was observed in 16 patients (73%). Interestingly, in one patient, a transformation from NSCLC to small-cell lung cancer (SCLC) was observed upon disease progression. Median PFS, since the start of osimertinib treatment, was 8.9 [interquartile range (IQR): 4.6–18.0] months, whereas median OS, since osimertinib initiation, was 20.7 (IQR: 8.8–27.7) months. Osimertinib was used as a second-line treatment in 11 (50%) patients, while 11 (50%) patients had received two or more lines of treatment prior to that with osimertinib. As expected, the latter group of patients had a significantly poorer outcome in terms of PFS and OS than the former (P<0.004 and 0.020, respectively). Clinical objective response rates (RECIST criteria) were observed in 14 (64%) patients. Oligoprogressive disease (oligo-PD) was noted in 9 (41%) patients, and in 7 of whom (78%) osimertinib was maintained for a median of 3.8 (IQR: 1.2–9.1) months beyond oligo-PD.
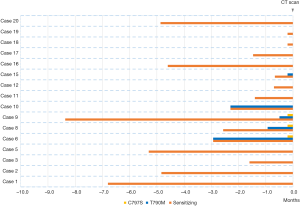
Median treatment durations of sequential gefitinib + osimertinib, afatinib + osimertinib and erlotinib + osimertinib were 30.1, 24.6 and 21.1 months, respectively, indicating that time on targeted therapy was longest in patients treated with the combination gefitinib + osimertinib combination. However, no significant differences were observed in OS and PFS according to first TKI treatment (afatinib, gefitinib, erlotinib). Figure 1 shows the times on targeted therapy and the time under osimertinib treatment for each patient.
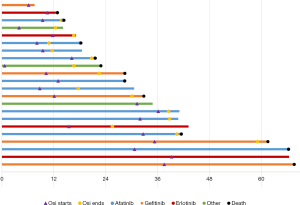
Considering toxicity, 12 patients reported adverse events, 82.6% of which were mild (G1). The most frequent toxicities were neutropenia (9%), diarrhea (9%), hypertransaminasemia (9%) and asthenia (9%). Only one G3 event was recorded (asymptomatic hyperamylasemia).
Longitudinal ctDNA monitoring
To analyze the evolution of these tumors throughout the course of treatment, EGFR somatic mutations within ctDNA were prospectively collected from 326 samples and analyzed by dPCR. A blood sample obtained before starting osimertinib treatment was available for all patients. At baseline, the p.T790M mutation was detected in 19 (86%) patients, with a median AF of 4.11% (minimum 0.1%; maximum 37.7%). In the other three cases, the p.T790M mutation was detected only in the re-biopsy (N=2) and in the cerebrospinal fluid (N=1). Noteworthy, two of these plasma-negative T790M patients each had metastases exclusively at the brain level. The original EGFR-sensitizing mutation was detected in all pre-treatment samples. Neither p.T790M AF nor the original EGFR-sensitizing mutation AF at the start of treatment predicted a survival benefit from osimertinib. Nevertheless, ctDNA levels across serial plasma samples were correlated with treatment responses. Specifically, undetectable levels of the original EGFR-sensitizing mutation after 3 months of osimertinib treatment were associated with improved PFS (HR: 0.19, 95% CI: 0.05–0.7). Similarly, patients in whom plasma levels of the original EGFR-sensitizing decreased after 3 months had a better prognosis in terms of PFS (HR: 0.14, 95% CI: 0.23–0.86). On the other hand, re-emergence of the original EGFR mutation, alone or together with the p.T790M mutation, was significantly associated with shorter PFS (HR: 8.8, 95% CI: 1.1–70.7 and HR: 5.9, 95% CI: 1.2–27.9, respectively), indicating that ctDNA quantification is informative in terms of prognosis also in this group of patients.
Molecular patterns upon disease progression
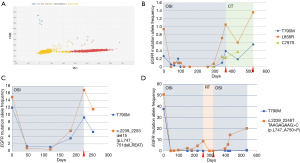
In order to assess the frequency of the p.C797S (c.2389T>A and c.2390G>C) mutation at the time of osimertinib progression in our population, dPCR was performed in all samples collected at osimertinib progression (N=16) (Figure 2A). At this time, the p.C797S mutation was found along with the p.T790M mutation as well as the original EGFR-sensitizing mutation in 3 (19%) patients (two cases with the p.L858R mutation and one with a deletion in exon 19). Specifically, two cases harbored the c.2390 G>C mutation and one featured the c.2389T>A mutation. Remarkably, dPCR analysis did not identify the p.C797S mutation in any of the previously collected samples, indicating that cells with this mutation were positively selected over the course of therapy. The p.C797S mutation was detected at a lower AF than p.T790M mutation levels, which, at the same time, were lower than the sensitizing mutation AF (Figure 2B). Interestingly enough, patients showing this “triplet pattern” (sensitizing+/T790M+/C797S+) tended to exhibit longer PFS and OS than patients who did not (P=0.1, Figure S1). In 2 patients (12.5%), plasma levels of the original EGFR-sensitizing mutation were again detected at the time of disease progression alongside the p.T790M mutation (Figure 2C). This “duplet pattern” (sensitizing+/T790M+) was detected in patients with a high tumor load. Finally, in the other 11 (69%) cases, there was a prominent increase in the original EGFR-sensitizing mutation, with null or residual levels of the p.T790M mutation detected (Figure 2D), suggesting that osimertinib was able to eliminate the p.T790M-mutated clone in this subset of patients (sensitizing+), even though the tumor was able to become resistant to treatment. The median time to progression in patients showing the triplet pattern (sensitizing+/T790M+/C797S+) was 12.27, 4.87 months in patients in whom only the original EGFR-sensitizing mutation was detected, and 2.17 months in patients with the duplet pattern (sensitizing+/T790M+). Figure S2 shows how early the appearance of the resistance mechanism was detected during ctDNA monitoring.
Next-generation sequencing (NGS) analysis upon osimertinib progression
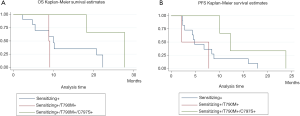
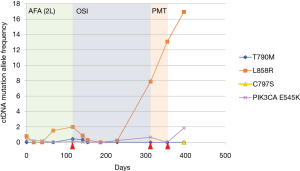
ctDNA collected at the time of disease progression was available from seven patients for NGS analysis. In this subset of patients, PIK3CA mutations were the alterations most frequently detected upon disease progression, being found in four patients. Specifically, we identified the p.E545K mutation in one patient (Table 2). The analysis of previous plasma samples by dPCR revealed that this mutation was not present at the start of the treatment (Figure 3). Likewise, the mutation p.E545A was detected at disease progression in three patients. Curiously, we detected the p.S464L mutation in the EGFR gene in a patient who was treated with cetuximab plus afatinib prior to osimertinib therapy. In addition, the p.A750P mutation in the EGFR gene was found in another patient who harbored the deletion in exon 19 p.L747_A750>P. Retrospective analysis of plasma samples revealed that the A750P mutation was also present at the start of osimertinib treatment although at a very low AF. Finally, an EGFR copy-number gain was detected by NGS in one case. However, this alteration could not be confirmed by any other alternative technique.

Full table
On the other hand, we found that the median TTD was 8.7 (IQR: 2.8–10.1) months in patients whose tumors harbored co-mutations in TP53, compared with 18 (IQR: 7.8–28.2) months in patients whose tumors were negative for TP53 mutations. However, the difference noted was not statistically significant, given the small sample size.
Discussion
There is growing evidence of the usefulness of liquid biopsy as an effective tool for biomarker testing and treatment monitoring. In the present study, the p.T790M mutation was detected in the plasma of 19 (86%) patients at baseline, supporting the clinical utility of liquid biopsies for decision-making about treatment. Nevertheless, the possibility of a false-negative result should be ruled out using tumor tissue obtained by biopsy (16). The reported sensitivities of the different assays for EGFR mutation detection using cfDNA from advanced NSCLC patients vary as much as from 30% to 100% (17). Although the cohort presented in this study is rather limited our results supports the usefulness of dPCR for plasma p.T790M testing. On the other hand, levels of the original EGFR-sensitizing mutation after 3 months of osimertinib treatment were of prognostic significance. Noteworthy, the effect size was substantive (HR: 0.19, 95% CI: 0.05–0.7). Several studies have reported that EGFR mutation tracking correlates with treatment outcome (11,12). However, it is important to mention that in the case of NSCLC patients resistant to first/second-generation EGFR-TKIs, treated with osimertinib, only the original EGFR-sensitizing mutation is informative for monitoring purposes. According to our data, a complete clearance of the p.T790M mutation was found in 69% of the patients with PD, and therefore, the p.T790M mutation is not useful in monitoring the response to osimertinib. In the same way, previous studies have reported similar results (18,19).
According to plasma genotyping, we were able to define three molecular patterns upon disease progression in patients treated with osimertinib, highlighting the importance heterogeneity in advanced disease. These patterns were also reported in a study cohort of 22 patients who became resistant to osimertinib and from whom cfDNA was collected during the phase I AURA study (7). Similarly, other studies have shown that the p.C797S mutation is always detected in conjunction with the p.T790M mutation as well as the original EGFR-sensitizing mutation (9,12,20). According to our data, these patterns may determine different prognoses. In our study, patients showing the “triplet pattern” (sensitizing+/T790M+/C797S+) tended to have better PFS and OS (P=0.1), suggesting that tumors that become resistant to osimertinib through p.T790M loss may have a poorer outcome. Likewise, Oxnard et al. reported that acquired resistance to osimertinib mediated by loss of the p.T790M mutation was associated with early treatment failure (21). However, despite its pertinence in this context, this observation requires confirmation in larger cohorts. NGS profiling of plasma samples has proved to be a valuable approach for identifying resistance mutations. In our hands, the activating mutations in codon 545 of the PIK3CA gene were frequently observed upon osimertinib progression. Likewise, other researchers have proposed that mutations in codon 545 of the PIK3CA gene constitute a common resistance mechanism of third-generation TKIs (22). Similarly, Yang et al. reported that mutations in PIK3CA potentially contribute to osimertinib resistance in patients without secondary EGFR mutations (23). In addition, we found the p.S464L mutation in the EGFR gene in the tumor of a patient treated with cetuximab plus afatinib prior to osimertinib therapy. Remarkably, this mutation has been reported in colorectal tumors that are refractory to cetuximab (24).
On the other hand, our results show that the efficacy of osimertinib in real-world practice was similar to that observed in clinical trials, with a favorable adverse effect profile. Similar results have recently been reported in a large sized real-world study (25). Strikingly, time on targeted therapy was longer in patients treated with the gefitinib + osimertinib combination, than those who received one of the other two combinations, although no significant difference in PFS according to first-line TKI was found.
It is important to mention that the small sample size of the present study is an important limitation and therefore although our results are of particular interest they need to be tested in appropriately sized cohorts.
Conclusions
In summary, we report a comprehensive descriptive study of a real-world cohort of patients treated with osimertinib as second-line treatment. Analysis of ctDNA during the course of the disease revealed three molecular patterns that might confer different prognoses. Besides the p.C797S mutation, putative PIK3CA mutations might underlie osimertinib resistance in patients without secondary EGFR mutations.
Acknowledgments
The authors wish to thank all the patients that participated in this study.
Funding: This study was supported by Carlos III Institute of Health, Spanish Ministry of Science and Innovation, and European Regional Development Fund (grant number: PI16/01818 and PI17/00064). E Sánchez-Herrero is supported by Plan de Empleo Juvenil Comunidad de Madrid (PEJ-2017-AI/SAL-6478), R Serna-Blasco is supported by Plan de Empleo Juvenil Comunidad de Madrid (PEJD-2018-PRE/BMD-8640), S Sanz-Moreno is supported by Plan de Empleo Juvenil Comunidad (PEJ-2017-TL/SAL-7476) and Alejandro Rodriguez-Festa is supported by Plan de Empleo Juvenil Comunidad (PEJ-2017-TL/SAL-7481).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/ tlcr.2020.04.01). AR reports other from Boehringer, Takeda during the conduct of the study. VC reports other from Roche, BMS, MSD, Pfizer, Lilly, AstraZeneca, Boehringer, Novartis, Takeda during the conduct of the study. MP reports other from Roche, BMS, MSD, Pfizer, Lilly, grants and other from AstraZeneca, Boehringer, other from Novartis, Takeda during the conduct of the study. MP serves as an unpaid editorial board member of Translational Lung Cancer Research from Sep 2019 to Sep 2021. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Hospital Puerta de Hierro Ethics Committee (approval number: internal code Acta nº02.16.). Written informed consent was obtained from the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Hirsch FR, Suda K, Wiens J, et al. New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet 2016;388:1012-24. [Crossref] [PubMed]
- Shih JY, Gow CH, Yang PC. EGFR mutation conferring primary resistance to gefitinib in non-small-cell lung cancer. N Engl J Med 2005;353:207-8. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015;21:560-2. [Crossref] [PubMed]
- Planchard D, Loriot Y, André F, et al. EGFR-independent mechanisms of acquired resistance to AZD9291 in EGFR T790M-positive NSCLC patients. Ann Oncol 2015;26:2073-8. [Crossref] [PubMed]
- Ou SI, Cui J, Schrock AB, et al. Emergence of novel and dominant acquired EGFR solvent-front mutations at Gly796 (G796S/R) together with C797S/R and L792F/H mutations in one EGFR (L858R/T790M) NSCLC patient who progressed on osimertinib. Lung Cancer 2017;108:228-31. [Crossref] [PubMed]
- Bersanelli M, Minari R, Bordi P, et al. L718Q mutation as new mechanism of acquired resistance to AZD9291 in EGFR-mutated NSCLC. J Thorac Oncol 2016;11:e121-3. [Crossref] [PubMed]
- Provencio M, Torrente M, Calvo V, et al. Dynamic circulating tumor DNA quantificaton for the individualization of non-small-cell lung cancer patients treatment. Oncotarget 2017;8:60291-8. [Crossref] [PubMed]
- Provencio M, Torrente M, Calvo V, et al. Prognostic value of quantitative ctDNA levels in non small cell lung cancer patients. Oncotarget 2017;9:488-94. [Crossref] [PubMed]
- Provencio M, Pérez-Barrios C, Barquin M, et al. Next-generation sequencing for tumor mutation quantification using liquid biopsies. Clin Chem Lab Med 2020;58:306-13. [Crossref] [PubMed]
- Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn 2017;19:4-23. [Crossref] [PubMed]
- Clark TG, Bradburn MJ, Love SB, et al. Survival analysis part I: basic concepts and first analyses. Br J Cancer 2003;89:232-8. [Crossref] [PubMed]
- Merker JD, Oxnard GR, Compton C, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 2018;36:1631-41. [Crossref] [PubMed]
- Mayo-de-Las-Casas C, Jordana-Ariza N, Garzón-Ibañez M, et al. Large scale, prospective screening of EGFR mutations in the blood of advanced NSCLC patients to guide treatment decisions. Ann Oncol 2017;28:2248-55. [Crossref] [PubMed]
- Del Re M, Bordi P, Rofi E, et al. The amount of activating EGFR mutations in circulating cell-free DNA is a marker to monitor osimertinib response. Br J Cancer 2018;119:1252-8. [Crossref] [PubMed]
- Del Re M, Rofi E, Cappelli C, et al. The increase in activating EGFR mutation in plasma is an early biomarker to monitor response to osimertinib: a case report. BMC Cancer 2019;19:410. [Crossref] [PubMed]
- Yu HA, Tian SK, Drilon AE, et al. Acquired resistance of EGFR-mutant lung cancer to a T790M-specific EGFR inhibitor: emergence of a third mutation (C797S) in the EGFR tyrosine kinase domain. JAMA Oncol 2015;1:982-4. [Crossref] [PubMed]
- Oxnard GR, Hu Y, Mileham KF, et al. Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M-positive lung cancer and acquired resistance to osimertinib. JAMA Oncol 2018;4:1527-34. [Crossref] [PubMed]
- Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:11815. [Crossref] [PubMed]
- Yang Z, Yang N, Ou Q, et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin Cancer Res 2018;24:3097-107. [Crossref] [PubMed]
- Arena S, Bellosillo B, Siravegna G, et al. Emergence of multiple EGFR extracellular mutations during cetuximab treatment in colorectal cancer. Clin Cancer Res 2015;21:2157-66. [Crossref] [PubMed]
- Marinis F, Wu YL, de Castro G Jr, et al. ASTRIS: a global real-world study of osimertinib in >3000 patients with EGFR T790M positive non-small-cell lung cancer. Future Oncol 2019;15:3003-14. [Crossref] [PubMed]


