
Top-level MET gene copy number gain defines a subtype of poorly differentiated pulmonary adenocarcinomas with poor prognosis
Introduction
Lung cancer is still the main cause for cancer related deaths worldwide. Understanding the mechanisms of molecular carcinogenesis of non-small cell lung cancer (NSCLC) is crucial to discover specific therapeutic targets and has led to improved outcome (1). However, although an increasing number of targeted therapies and immuno-oncology related treatments is available nowadays, NSCLC still remains a deadly disease since only a minority of patients can be cured (2). One of the biologically and therapeutically relevant targets in NSCLC and many other human cancers is the mesenchymal-epithelial transition receptor (MET) and its ligand, hepatocyte growth factor (HGF) (3,4). The MET proto-oncogene was initially described by Cooper et al. in an osteosarcoma derived cell line in 1984 (5). The MET gene is located on chromosome 7q and its product, a heterodimeric transmembrane receptor tyrosine kinase, consists of an extracellular α- and a transmembrane β-chain (1,3).
MET as a receptor tyrosine kinase can be activated by a multitude of biologic mechanisms, such as gene fusions, activating mutations, gene amplification and also simply by overexpression of the receptor protein or by ligand dependent activation. MET activation itself leads to dimerization and transphosphorylation followed by activation of downstream signaling via PI3K/AKT, RAS-RAC/RHO, MAPK and phospholipase C pathways (6). The effects are manifold: the MET/HGF pathway has an impact on multiple cellular functions, such as differentiation, cell cycle progression, proliferation and angiogenesis (7). Its dysregulation occurs in many different types of cancer (4) and leads to several effects in tumorigenesis, such as cancer cell proliferation, invasion, survival, motility and the development of metastases (8).
Activating missense mutations in the tyrosine kinase domain have been described in papillary renal cancer (9). Another type of activating MET mutations affects the splice site donor and acceptor regions around exon 14. Alternative splicing with consecutive skipping of exon 14 causes a stabilization and accumulation of catalytically active MET protein on the cell surface due to reduced ubiquitinylation and proteasomal degradation. Originally discovered in small cell lung cancer, MET exon 14 skipping mutations have also been described in 3–6% of adenocarcinoma of the lung and about 1–2% of tumors with other NSCLC histologies (10-14). Moreover, MET exon 14 skipping mutations were identified as an independent prognostic factor that predict poor survival (15,16). MET amplification has been described in about 3-5% of newly diagnosed NSCLC (15,17,18) and increased MET gene copy number seems to be a negative prognostic factor (17,19-21).
Many tyrosine kinase inhibitors with anti-MET activity are currently being explored in cancers with MET activation, among them MET amplified and mutated NSCLC. Early data from clinical trials is available mainly for crizotinib, capmatinib and tepotinib (22). Recently, Camidge et al. presented an update of the PROFILE 1001 study reporting on MET targeting therapy with crizotinib in 40 NSCLC patients (23). Those with high MET amplification [defined by MET/centromere 7 (CEP7) ratio ≥4] showed clinically meaningful antitumor activity with rapid and durable responses. Objective response rates were lower in tumors with lower MET amplification levels. Thus, based on available data, MET amplification is probably both, a negative prognostic and a potential predictive biomarker for MET tyrosine kinase inhibitors. However, generally accepted criteria for MET positivity in NSCLC do not yet exist. Moreover, even methods to detect clinically meaningful MET alterations are still under discussion. MET mutations, i.e. those mutations which cause exon 14 skipping, and gene fusions can be detected by DNA-based next generation sequencing of the intron-exon borders around exon 14 of the MET gene. Additionally, RNA-based approaches are employed. Also, gene copy number gains can be detected by some sequencing assays. However, fluorescence in situ hybridization (FISH) has been used to select patients with MET amplification in clinical trials on MET inhibitors so far (23,24). Detections of MET protein expression by immunohistochemistry (IHC) was shown to be associated with amplification to a certain extent (18). However, a clinical trial with the therapeutic monoclonal MET antibody onartuzumab failed to demonstrate a clinically meaningful predictive value of MET IHC (25,26). Based on currently available treatment approaches in NSCLC with MET inhibitors including clinical trials, two types of predictive biomarkers seem to be the most promising: (I) DNA or RNA sequencing for exon 14 skipping mutations, and (II) FISH for amplification.
However, various and different, sometimes even contradictory criteria for MET amplification or MET copy number gains have been proposed. Some authors have used a high MET/centromere 7 copy number ratio as a measurement for amplification (17,19,23). Since high level MET copy number gains can also occur against the background of simultaneously increased copies of centromeric regions (resulting in a “negative” ratio <2.0), we have previously suggested a more general approach to describe copy number changes of MET (18). This approach specifically emphasizes average gene copy number and has been adopted in current clinical trials (24).
Data on the frequency of different amplification levels in NSCLC patients are still sparse. Moreover, MET gene copy number gains have not been comprehensively correlated with clinical data so far. In this study, we aimed at elucidating the frequencies of MET amplification levels in an unbiased series of consecutive clinical samples of NSCLC patients and correlating these levels with different histologies, molecular subtypes and outcome of patients. Since effects of MET inhibitors seem to be related to a gene-dose effect at least to a certain degree, we furthermore aimed to establish the molecular subgroup of NSCLC with the highest unequivocal MET amplification level and to describe the histologic and clinical phenotype of this newly defined subgroup.
Methods
Patients
A total of 390 unselected consecutive NSCLC have been included in this study. Patients were tested for MET amplification between January 1st, 2015 and June 30th, 2017 as part of the routine molecular diagnostics at the Institute of Pathology of the University Medical Center Göttingen, Germany. Seventeen patients were excluded due to missing clinical data, resulting in a series of 373 consecutive NSCLC patients. Tumor stage was determined based on the 8th edition of UICC TNM Staging System of lung cancer (27). Cases which were initially staged on the basis of the 7th edition were double-checked and re-staged if appropriate. For subsequent data analysis, stages IVA and IVB were aggregated to stage IV and compared with lower stages. All patients were treated at a dedicated lung tumor center (Lungentumorzentrum Universität Göttingen). Clinical and follow-up data were obtained from their medical records.
All patients were treated according to local standards which are based on national and international guidelines, and if necessary, received systemic therapy. Treatment information was analyzed retrospectively in full detail for all patients with MET gene copy number gains (defined by MET level ≥1; see below; n=141). 70.2% (99/141) of these patients received systemic therapy at any time. First line therapy consisted of platinum-based combination treatments (n=90 patients, with pemetrexed, paclitaxel, docetaxel, gemcitabine, or etoposide). In 22 of these patients, a triple combination with bevacizumab was given. In patients who received definitive radio-chemotherapy, cisplatin was combined with vinorelbine or given as a monotherapy. In addition, single agent systemic therapies in stage IV disease included pemetrexed, and erlotinib or afatinib in EGFR mutant cancers. 48.5% (48/99) of patients were treated in a 2nd line setting who received docetaxel with or without nintedanib, platinum-based combinations with paclitaxel, vinorelbine, or etoposide, triple combinations with bevacizumab, or a monotherapy with pemetrexed, afatinib, erlotinib, or gefitinib. 11 patients were treated with PD-L1 or PD-1 inhibitors (atezolizumab, nivolumab or pembrolizumab in 1, 7 and 3 patients, respectively), and 7 patients received an anti-MET tyrosine kinase inhibitor (TKI) as 2nd line treatment. 3rd line therapy was given in 21/99 patients (21.2%; nivolumab: n=11; MET TKI: n=1, further therapies included erlotinib, pemetrexed, carboplatin plus paclitaxel, and docetaxel with or without ramucirumab).
Only a small number of patients (9/99, 9.1%) was also treated in a 4th line setting. These therapies were based on atezolizumab in one patient and nivolumab in another two patients, furthermore docetaxel with or without ramucirumab, gemcitabine, vinorelbine, and erlotinib. Two patients were treated with docetaxel in a 5th line setting, one with further vinorelbine at 6th line and with nivolumab at 7th line.
This study was conducted after approval of the local Ethics committee (5/1/17).
Histology, subtyping and molecular profiling
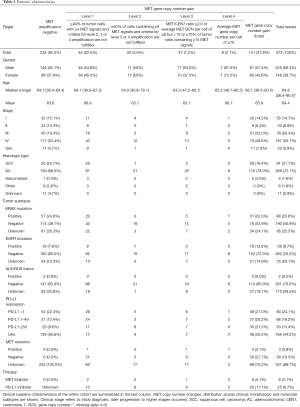
The majority of the specimens (60%) consisted of primary tumor tissue, predominantly transbronchial biopsies followed by core needle biopsies and resection specimens, and one third were surgical resections and biopsies of distant metastases. Cytology specimens (i.e., smears or cytospin preparations) were excluded. Tumor diagnoses were established based on the current WHO and IASLC classification (28,29). In brief, tumors with strong and predominant expression of p40 were classified as squamous cell carcinomas; lesions with convincing expression of neuroendocrine markers, e.g., CD56, chromogranin A, synaptophysin, were regarded as neuroendocrine tumors; p40-negative non-neuroendocrine carcinomas, either gland forming or solid, with or without TTF-1 expression were recognized as adenocarcinomas. Additional diagnostic biomarkers were applied if appropriate. For further statistical analyses tumors were grouped into four subgroups: (I) adenocarcinomas, (II) squamous cell carcinomas, (III) sarcomatoid carcinomas (comprising pure spindled or pleomorphic carcinomas), (IV) others (include large cell neuroendocrine carcinomas, typical/atypical carcinoids). Frequencies of tumor subtypes are summarized in Table 1.
Full table
All non-squamous NSCLC cases underwent further molecular characterization. ALK and ROS1 testing was done by FISH as previously described (30-32). For EGFR and KRAS sequencing, the EGFR and KRAS therascreen assays, (Qiagen, Hilden, Germany) were applied according to the manufacturer’s recommendations. Next generation sequencing was done as previously described (33). All cases were further tested for PD-L1 expression by using the clone 28-8 on a DAKO Omnis platform (staining protocol described in detail in Koppel et al. 2018 (34); evaluation and scoring is described in Schildhaus et al. 2015 (18).
MET FISH
FISH has been carried out as previously described (18,35). In brief, 4 µm thick sections have been hybridized by using the ZytoLight SPEC MET/CEN7 Dual Color Probe (ZytoVision, Bremerhaven, Germany). The numbers of MET and centromere 7 signals were counted in 60 nuclei obtained from three different areas with the highest gene count. Average gene copy number and ratio (MET/CEN7) as well as the percentages of tumor cells with ≥4, ≥5 and ≥15 gene copies were calculated for each tumor. All FISH assays were evaluated by two pathologists with specific experience in this field (KS, HUS). Tumors were categorized into amplification levels, based on previously published criteria after modification (see below, Tables 1,2).

Full table
Statistics
SPSS software (IBM Corp., IBM SPSS Statistics for Windows, Version 25.0. Armonk, NY USA) was used for statistical analysis. Categorial variables were tested by chi-square or Fisher’s exact test. Survival times were analyzed with the Kaplan-Meier method and for comparing the survival times across different groups, the log rank test was applied. Survival data were available for 371 patients. Overall survival (OS) was calculated based on the date of first diagnosis to the date of last follow up or death of patient. Patients who were still alive or were lost to follow-up were censored at the time of the last contact. Cox regression was used to assess univariate tests and multivariate models. Parameters with P values <0.1 at Cox regression in univariate analysis were tested in multivariate models. Wald test was performed testing different subtypes of categorical variables. Forest plot graph was performed using GraphPad Prism software (GraphPad Software, GraphPad Prism for Windows, version 8.0.1, La Jolla, California USA). All tests were two-sided and statistical significance was defined as P<0.05.
Results
Frequencies of MET gene copy number alterations and definition of a top-level amplification category
Two hundred thirty-two samples (62.2%) were MET amplification negative and 141 showed MET gene copy number gains at various levels. Distribution of MET copy number changes across clinical, morphologic and molecular subgroups together with baseline characteristics of the entire cohort are shown in Table 1. Five out of six pure sarcomatoid carcinomas showed MET gene copy number (GCN) gains (Table 1).
Average MET gene copy numbers per nucleus ranged from 2.4 to a maximum of 25.2 (median 4.4). MET/CEN 7 ratios were found between 0.8 and 10.1 (median: 1.4). To determine the patients’ subgroup with the highest unequivocal MET amplification level, we determined that parameter with the broadest numerical range (i.e., average gene copy number) and calculated the 90th percentile which was found at 10.8 MET gene copies per tumor cell. Therefore, a top-level MET amplification category (level 4) was defined by an average gene copy number of ≥10 MET signals per nucleus (Table 2). MET positivity was found in 84, 22, 27 and 8 patients at levels 1, 2, 3 and 4, respectively (Tables 1,2). Among level 4 samples, MET/CEN 7 ratio ranged from 1.4 to 10.1. Four out of eight MET top-level patients had a MET/CEN7 ratio ≥4.0; the remaining cases showed co-amplification of centromeric sequences (CEN7). However, a high ratio ≥4.0 was also found in two patients with less MET gene copies at lower amplification levels. MET GCN gain—at any level—and ALK or ROS1 gene fusions did not co-occur. Eighteen patients (50%) with an activating EGFR mutation showed a simultaneous MET amplification which was detectable prior to EGFR TKI treatment. Three (2.1%) patients were tested positive for MET exon 14 skipping mutation and were classified with a level 1, 3 and 4 MET GCN gain. Two of those patients were initially diagnosed with stage IV and one with stage III.
Clinical and morphologic phenotype of patients with top-level MET amplification (level 4)
We identified eight MET top-level patients, seven of them were male (87.5%). Predominance of male gender was the highest among these patients if compared with all other MET status. Median age was 65.2 years (range, 48.7–90.5 years). Most of the patients were diagnosed with multiple metastases; five patients were initially with stage IV, two with stage III and one with stage I at the time of initial diagnosis. Many level 4 patients had metastatic lesions at uncommon locations, such as skin, muscle or pararectal soft tissue. We obtained information about the smoking status for 6 out of 8 patients. All of them were heavy smokers with a history of at least 40 pack years.
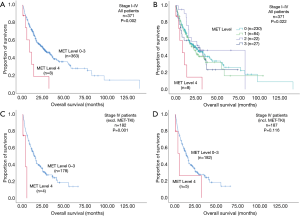
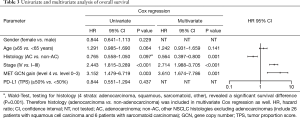
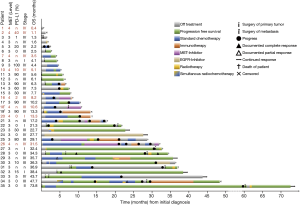
Patients with MET top-level amplifications survived significantly shorter than all other patients in our series [median overall survival (mOS) 8.2 vs. 23.6 months; P=0.002, Log Rank test; Figure 1]. Uni- and multivariate analysis demonstrated stage, histotype and MET amplification level as the only independent parameters for outcome. It is noteworthy that adenocarcinomas per se are significantly associated with better outcome than all other NSCLC histologies. This effect, however, is overcompensated by top-level MET amplification which occurred exclusively in adenocarcinomas. Level 4 is associated with a more than threefold increase of the likelihood to die of the cancer (hazard ratio: 3.61; Table 3) which is independent of the clinical stage at initial presentation. Survival of patients with top-level MET amplification (level 4) was shorter even if compared with high level amplified tumors (level 3) and even if systemic treatment was administered in a multitude of therapy lines. Figure 2 demonstrates the treatments applied to MET level 3 and 4 patients, and visualizes the individual progress of disease at different levels of MET amplification. Off label treatment with a MET tyrosine kinase inhibitor was beneficial in one level 4 patient and led to a partial response.
Full table
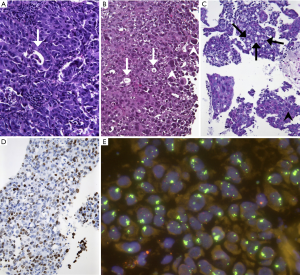
Also, in terms of morphology, we observed a peculiar phenotype: All MET level 4 patients were basically classified as adenocarcinoma. No squamous cell carcinoma was found among these patients (excluded by lack of p40 expression in all samples). All of these cancers were poorly differentiated, predominantly solid adenocarcinomas with pleomorphic features in terms of interspersed pleomorphic giant cells. A pure or predominant pleomorphic or sarcomatoid differentiation, however, was not seen. Interestingly, five out of eight lesions showed TTF1-positivity which was, however, heterogeneous and moderate (Figure 3). Regarding molecular subgroups, neither EGFR mutation nor ALK/ROS1 fusions were found among MET level 4 patients, but one KRAS mutation and one MET exon 14 skipping mutation were found. High PD-L1 expression (TPS ≥50%) could not be demonstrated in these tumors.
Discussion
In this study we describe a novel subgroup of NSCLC patients which is defined by the highest unequivocal level of MET amplification. We examined a prospective series of unselected consecutive NSCLC samples by FISH. Based on a comprehensive descriptive approach, we defined the cut-off numerically at the 90th percentile of the average MET gene copy number per tumor cell. Among all tested parameters, i.e., MET/CEN 7 ratio, average gene copy number and percentages of tumor cells with ≥4, ≥5 and ≥15 MET gene copies per tumor cell, average MET gene copy number showed the broadest numerical range. Therefore, the MET top-level amplification category was defined by ≥10 MET gene copies per tumor cell on average.
Furthermore, we correlated cases fulfilling our newly described criteria with clinical and morphologic data and demonstrate a peculiar phenotype of these patients. A major finding is related to clinical outcome: Among all characteristics tested—including stage, histotype and molecular subtypes—patients with MET top-level amplification suffer from the shortest survival and the highest likelihood to die from their cancer. Thus, MET top-level amplification is an independent prognostic factor which is even unrelated to clinical stage at initial diagnosis. In our series the prevalence of MET top-level amplification which was 2.1%. This is basically in line with a previous report (18). In a retrospective re-calculation of data from our former publication the prevalence of this subgroup was 0.9%. Combined data (based on 1,066 prospectively and comparably tested patients from that publication and from this study), therefore, indicate that the prevalence of MET top-level amplification is in the range of 1% to 2% among Caucasian patients from Western countries. Furthermore, we could demonstrate that MET top-level amplification is mutually exclusive with EGFR mutations as well as with ALK and ROS1 gene fusions. However, activating KRAS mutations and MET exon 14 skipping mutations can co-occur whereas high PD-L1 expression has not been found in this subgroup.
MET top-level amplified lung cancers seem to show also a specific morphologic phenotype which we describe as poorly differentiated adenocarcinomas with pleomorphic features. We acknowledge that MET top-level samples show some similarities with pleomorphic and/or sarcomatoid carcinomas where MET alterations, including MET mutations and lower levels of amplification, have been already described. However, adenocarcinoma-typic features such as gland formation TTF1 expression and/or KRAS mutation could be demonstrated in our cases which fulfilled the criteria for top-level amplification. Therefore, we feel encouraged to describe our cases as examples of a specific subtype of pulmonary adenocarcinomas rather than sarcomatoid carcinomas. In addition and supporting our recent finding, all patients with top-level amplification from that previous report turned out retrospectively as adenocarcinomas (18).
We are aware that the small size of our MET top-level cohort is a major limitation of our study. However, we provide here first evidence that a peculiar subgroup of NSCLC exists which is characterized by the highest unequivocal MET amplification and a specific morphologic and clinical phenotype. This observation may contribute to a more specific description of “MET amplification” which has been subject to rather vague definitions in the past.
One major finding of our study was the extremely poor prognosis of these MET top-level amplified cancers which constituted the worst prognostic subgroup of all NSCLC histologies in our cohort. This observation was not only unrelated to initial clinical stage but also nearly independent of any form of systemic treatment, including conventional chemotherapy and immune-checkpoint inhibitor treatment. Therefore, we conclude that MET blockade might become a reasonable systemic treatment option for these patients. Although we cannot provide systematic response data of MET inhibitors from our prevalence study, we believe that the newly defined MET top-level amplification may provide a reasonable inclusion criterion for ongoing clinical trials with MET inhibitors in NSCLC. Based on the extremely short survival of these patients, we suggest to give patients with MET top-level amplification early access to anti-MET treatment already in a first line approach. Otherwise, the aggressive biology of these tumors may overcompensate a potential benefit of MET blockade if patients are treated too late. Our limited clinical experience with MET inhibitors obtained in this study may support the hypothesis that early treatment of patients with top-level MET amplification may benefit from treatment with a MET TKI. This, however, needs to be proven by prospective clinical trials. Based on our data, we suggest (I) to establish prospective randomized trials enrolling patients with MET top-level amplified cancers in a first line setting, and (II) to re-analyze subgroups of top-level amplified NSCLC patients from ongoing or terminated trials.
The definition of “MET positivity” is still under debate. Whereas MET mutations are commonly accepted as an actionable target, it is still unclear whether MET amplification may also be actionable. Very recently, Camidge et al. presented an update of the PROFILE 1001 study reporting on MET targeting therapy with crizotinib in 40 NSCLC patients. Patients with high MET amplification [in this study defined by MET/centromere 7 (CEP7) ratio ≥4] showed clinically meaningful antitumor activity with rapid and durable responses. Objective response rates for low level MET/CEP7 ratio (1.8–2.1, n=1), medium level (2.2–3.9, n=14) and high level (≥4, n=20) were 33.3%, 14.3%, and 40.0%, respectively. Best median progression free survival was detected in the high level group (6.7 months), 1.8 and 1.9 months for low and medium level, respectively (23). This observation may point towards a gene dose effect which may be meaningful for a significant effect of these drugs. Moreover, we conclude that only patients with the highest unequivocal MET amplification level may be good candidates for anti-MET treatment. In this context, we need to emphasize that low level MET copy number gains have probably no specific value as a prognostic or predictive biomarker. Many cancers do show slight or moderate increases of MET copy numbers which do not necessarily reflect a specific biologic mechanism in terms of an oncogenic driver.
In summary, we describe a subtype of NSCLC which can be determined by very high MET gene copy number gains (i.e., top-level amplification as defined by ≥10 MET gene copies per tumor cell on average). We provide first evidence that tumors with this particular feature account for 1% to 2% of NSCLC cases and share a common clinical, genetic and morphologic phenotype. Patients with MET top-level cancers suffer from a deadly and aggressive tumor with extremely short overall survival which does not adequately respond to conventional chemotherapy or immune-therapy. MET top-level amplification is mutually exclusive with actionable EGFR, ALK or ROS1 alterations, whereas KRAS and MET mutations may co-occur. These particular tumors show a characteristic morphologic phenotype describable as adenocarcinoma with pleomorphic features.
Preliminary data from clinical trials with MET inhibitors point toward a gene dose effect. Therefore, we suggest including patients with MET top-level amplification specifically in clinical trials, also in first line settings.
Acknowledgments
The authors are grateful to the expert technical assistance provided by Mercedes Martin-Ortega, Sina Eckstein, Meike Wendt, Judit Wolf-Salgo, Anke Klages and Nicole Kerl.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-19-339). TRO reports personal fees from AstraZeneca, personal fees from BMS, personal fees from Boehringer-Ingelheim, personal fees from Eli Lilly, personal fees from Medac, personal fees from MSD, personal fees from Novartis, personal fees from Roche/Genentec, personal fees from Sanofi-Aventis, outside the submitted work. KS reports personal fees from MSD Germany, personal fees and non-financial support from Roche Austria, personal fees and non-financial support from Novartis Austria, outside the submitted work. AR reports grants from AbbVie, grants from AstraZeneca, grants from BMS, grants from Boehringer Ingelheim, grants from Eli Lilly, grants from MSD, grants from Novartis, grants from Pfizer, grants from Roche, outside the submitted work. HUS reports grants and personal fees from Novartis Oncology, personal fees from MSD, personal fees from BMS, personal fees from Pfizer, personal fees from ZytoVision, personal fees from Roche, from Abbvie, personal fees from Zytomed Systems, outside the submitted work. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was conducted after approval of the local Ethics committee (5/1/17) and informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Robinson KW, Sandler AB. The Role of MET Receptor Tyrosine Kinase in Non-Small Cell Lung Cancer and Clinical Development of Targeted Anti-MET Agents. Oncologist 2013;18:115-22. [Crossref] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Organ SL, Tsao M-S. An overview of the c-MET signaling pathway. Ther Adv Med Oncol 2011;3:S7-19. [Crossref] [PubMed]
- Sadiq AA, Salgia R. MET As a Possible Target for Non–Small-Cell Lung Cancer. J Clin Oncol 2013;31:1089-96. [Crossref] [PubMed]
- Cooper CS, Park M, Blair DG, et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature 1984;311:29-33. [Crossref] [PubMed]
- Cecchi F, Rabe DC, Bottaro DP. Targeting the HGF/MET signalling pathway in cancer therapy. Expert Opin Ther Targets 2012;16:553-72. [Crossref] [PubMed]
- Blumenschein GR, Mills GB, Gonzalez-Angulo AM. Targeting the Hepatocyte Growth Factor–cMET Axis in Cancer Therapy. J Clin Oncol 2012;30:3287-96. [Crossref] [PubMed]
- Birchmeier C, Birchmeier W, Gherardi E, et al. Met, metastasis, motility and more. Nat Rev Mol Cell Biol 2003;4:915-25. [Crossref] [PubMed]
- Schmidt L, Duh FM, Chen F, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet 1997;16:68-73. [Crossref] [PubMed]
- Ma PC, Kijima T, Maulik G, et al. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res 2003;63:6272-81. [PubMed]
- Ma PC, Jagadeeswaran R, Jagadeesh S, et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res 2005;65:1479-88. [Crossref] [PubMed]
- Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov 2015;5:850-9. [Crossref] [PubMed]
- Paik PK, Drilon A, Fan PD, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov 2015;5:842-9. [Crossref] [PubMed]
- Heist RS, Shim HS, Gingipally S, et al. MET Exon 14 Skipping in Non-Small Cell Lung Cancer. Oncologist 2016;21:481-6. [Crossref] [PubMed]
- Tong JH, Yeung SF, Chan AWH, et al. MET Amplification and Exon 14 Splice Site Mutation Define Unique Molecular Subgroups of Non-Small Cell Lung Carcinoma with Poor Prognosis. Clin Cancer Res 2016;22:3048-56. [Crossref] [PubMed]
- Yeung SF, Tong JHM, Law PPW, et al. Profiling of Oncogenic Driver Events in Lung Adenocarcinoma Revealed MET Mutation as Independent Prognostic Factor. J Thorac Oncol 2015;10:1292-300. [Crossref] [PubMed]
- Cappuzzo F, Marchetti A, Skokan M, et al. Increased MET Gene Copy Number Negatively Affects Survival of Surgically Resected Non–Small-Cell Lung Cancer Patients. J Clin Oncol 2009;27:1667-74. [Crossref] [PubMed]
- Schildhaus HU, Schultheis AM, Rüschoff J, et al. MET Amplification Status in Therapy-Naïve Adeno- and Squamous Cell Carcinomas of the Lung. Clin Cancer Res 2015;21:907-15. [Crossref] [PubMed]
- Go H, Jeon YK, Park HJ, et al. High MET Gene Copy Number Leads to Shorter Survival in Patients with Non-small Cell Lung Cancer. J Thorac Oncol 2010;5:305-13. [Crossref] [PubMed]
- Guo B, Cen H, Tan X, et al. Prognostic value of MET gene copy number and protein expression in patients with surgically resected non-small cell lung cancer: a meta-analysis of published literatures. PLoS One 2014;9:e99399. [Crossref] [PubMed]
- Dimou A, Non L, Chae YK, et al. MET Gene Copy Number Predicts Worse Overall Survival in Patients with Non-Small Cell Lung Cancer (NSCLC); A Systematic Review and Meta-Analysis. PLoS One 2014;9:e107677. [Crossref] [PubMed]
- Drilon A, Cappuzzo F, Ou SI, et al. Targeting MET in Lung Cancer: Will Expectations Finally Be MET? J Thorac Oncol 2017;12:15-26. [Crossref] [PubMed]
- Camidge DR, Otterson GA, Clark JW, et al. Crizotinib in patients (pts) with MET-amplified non-small cell lung cancer (NSCLC): Updated safety and efficacy findings from a phase 1 trial. J Clin Oncol 2018;36:9062. [Crossref]
- Novartis Pharmaceuticals. Clinical Study of Oral cMET Inhibitor INC280 in Adult Patients With EGFR Wild-type Advanced Non-small Cell Lung Cancer. ClinicalTrials.gov. 2018. Available online: https://clinicaltrials.gov/ct2/show/NCT02414139
- Spigel DR, Edelman MJ, O’Byrne K, et al. Results From the Phase III Randomized Trial of Onartuzumab Plus Erlotinib Versus Erlotinib in Previously Treated Stage IIIB or IV Non–Small-Cell Lung Cancer: METLung. J Clin Oncol 2017;35:412-20. [Crossref] [PubMed]
- Brower V. Onartuzumab ineffective in non-small-cell lung cancer. Lancet Oncol 2017;18:e66. [Crossref] [PubMed]
- Brierley JD, Gospodarowicz MK, Wittekind C. editors. Classification of Malignant Tumours. 8th ed. John Wiley & Sons, 2017
- Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Classification of Lung Adenocarcinoma. J Thorac Oncol 2011;6:244-85. [Crossref] [PubMed]
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Bos M, Gardizi M, Schildhaus HU, et al. Complete metabolic response in a patient with repeatedly relapsed non-small cell lung cancer harboring ROS1 gene rearrangement after treatment with crizotinib. Lung Cancer 2013;81:142-3. [Crossref] [PubMed]
- Schildhaus HU, Deml KF, Schmitz K, et al. Chromogenic in situ hybridization is a reliable assay for detection of ALK rearrangements in adenocarcinomas of the lung. Mod Pathol 2013;26:1468-77. [Crossref] [PubMed]
- Overbeck TR, Schmitz K, Engelke C, et al. Partial Response to First-Line Crizotinib in an Elderly Male Patient with ROS1 Translocation-Positive Lung Cancer. Case Rep Oncol 2016;9:158-63. [Crossref] [PubMed]
- Chen NM, Neesse A, Dyck ML, et al. Context-Dependent Epigenetic Regulation of Nuclear Factor of Activated T Cells 1 in Pancreatic Plasticity. Gastroenterology 2017;152:1507-1520.e15. [Crossref] [PubMed]
- Koppel C, Schwellenbach H, Zielinski D, et al. Optimization and validation of PD-L1 immunohistochemistry staining protocols using the antibody clone 28-8 on different staining platforms. Mod Pathol 2018;31:1630-44. [Crossref] [PubMed]
- Schmitz K, Koeppen H, Binot E, et al. MET Gene Copy Number Alterations and Expression of MET and Hepatocyte Growth Factor Are Potential Biomarkers in Angiosarcomas and Undifferentiated Pleomorphic Sarcomas. PLoS One 2015;10:e0120079. [Crossref] [PubMed]


