cfDNA changes for monitoring of targeted therapy in a primary EGFR mutation lung adenocarcinoma
EGFR-TKI treatments are recommended to be used in non-small cell lung carcinomas with sensitizing mutations, and acquired resistance still is a vital issue for the treatment. More than 50% of resistance develops EGFR T790M (1-3). Besides, MET amplification has been described as a resistance mechanism in 5% to 20% of patients treated with first/second generation and up to 30% with third-generation (osimertinib) TKIs (4). Here We report a rare case of later T790M mutation that occurred with a secondary MET amplification after first line icotinib treatment in EGFR-mutated lung adenocarcinoma. The emergence of two resistance mechanisms after EGFR-TKI may be challenging for successful treatment.
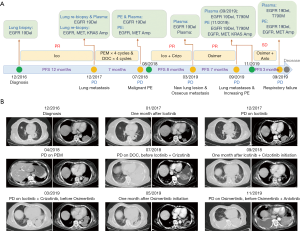
The patient was a 65-year-old female never-smoker who presented with intermittent chest pain and was found to have bilateral lung infiltrates with a maximum of a 6×5 cm left lower lobe mass and mediastinal adenopathy. CT-guided lung biopsy showed lung adenocarcinoma positive for an EGFR exon 19del mutation by direct sequencing. Then she was started on induction icotinib (150 mg tid PO). Image scans after four weeks showed partial response (PR) of the lung mass. After a progression-free survival (PFS) of ~12 months, she experienced progressive disease (PD) with new lung metastasis. Liquid biopsy and CT-guided lung re-biopsy revealed newly acquired MET amplification (2-fold in Tissue) conjunction with EGFR 19del (level: plasma 20.2%; tissue 87%), while no T790M mutation by next generation sequencing (NGS) assay. NGS also detected TP53 mutation (level: plasma 1.7%; tissue 32.8%), and amplification of EGFR (6.3-fold in tissue) and KRAS (2.1-fold in tissue). She was switched to palliative chemotherapy with single pemetrexed due to a patient’s score of ECOG performance status is two and fear of chemotherapy. Unfortunately, she developed progression as she was completing four cycles of chemotherapy, and then switched to third-line chemotherapy with single docetaxel. Also, she was undergoing disease progression with a left lung and mediastinal adenopathy, accompany with new malignant pleural effusions (PE). Molecular analysis by NGS was again performed and revealed persistent EGFR 19del (level: Plasma 27.9%; PE 84.8%), TP53 mutation (level: Plasma 1.9%; PE 74.3%), MET and EGFR amplification (2.9- and 3.9-fold in PE). The patient’s clinical condition rapidly switched to a performance status of 4 when coming back in 2018-8. The patient developed respiratory failure with assisted oxygen requirement, and fourth-line joint therapy with crizotinib (200 mg bid PO) and icotinib (150 mg tid PO) was urgently initiated. The patient’s symptoms significantly improved within three days. Restaging scans obtained after one month of crizotinib and icotinib joint therapy showed marked pulmonary improvement and resolution of PE. Seven months after treatment, she developed new chest wall pain and dyspnea. Repeat scans showed multiple new lung lesions and progressive osseous metastases. Plasma NGS detected original EGFR 19del (level: plasma 15.1%), TP53 (level: plasma 0.9) and newly acquired EGFR T790M (level: plasma 2.6%). Then, the patient was then treated with osimertinib and experienced significant improvement in dyspnea and pain, and CT scan after 1-month osimertinib therapy showed a dramatic tumor response. However, after approximately seven months of therapy, the patient was admitted to the hospital with further dyspnea and malaise. Imaging showed progressive lung metastases and increasing pleural effusion. NGS revealed variational EGFR 19del (level: PE 76.6%), EGFR T790M (level: PE 0.1%), TP53 mutation (level: 93.9%), MET and EGFR amplification (2.9- and 6.0-fold in PE). Then, the patient was suggested to take Anlotinib, an antivascular drug, combined with osimertinib, and she experienced ~three months of stable disease (SD) until 2020-1. The patient developed worsening respiratory failure with continuously assisted 3 L/min oxygen requirement when returned to the clinic in 2020-1, re-test by NGS revealed persistence of EGFR19del (level Plasma 20.5%; PE 66%), EGFR T790M (level Plasma 0.7%; PE not detected), TP53 mutation (level Plasma 2.5%; PE 70%), MET and EGFR amplification (4.0- and 3.0-fold in PE) (Figure 1). The patient clinical condition rapidly worsened due to respiratory failure, and she died a few days later.

EGFR T790M accounts for more than 50% mechanism of first-generation TKI resistance, MET amplification accounts for 5% to 20% resistance mechanism in patients treated with first generation. Generally speaking, EGFR T790M mutations are firstly reported when drug resistance occurs after first-generation EGFR-TKI (icotinib), followed by MET amplification after resistance to third-generation (osimertinib) in NSCLC EGFR-mutated patient. What’ more, several studies report MET amplification, sometimes co-occur with EGFR T790M as molecular mechanisms of resistance to EGFR TKIs (5,6). However, we are reporting a rare case of a patient with EGFR-mutated lung adenocarcinoma who had an excellent initial response to icotinib, with a MET amplification detected by liquid biopsy when first acquired resistance occurred, and the patient showed clear clinical and radiographic response to the combination therapy with crizotinib and icotinib, followed by progression, at which time plasma cell-free DNA analysis identified acquired EGFR T790M mutation that was not detected at the time of the first acquired resistance diagnosis. After ~seven months of PFS of osimertinib treatment, the patient developed drug-resistance, meantime liquid biopsy analysis identified clearance of EGFR T790M mutation and reappearance of MET amplification in liquid (PE) biopsy (Figure 2). Efforts have been made to interrogate mechanism of targeted therapy in a primary EGFR mutation lung adenocarcinoma. Analysis of EGFR on DNA from MET amplification clones revealed that both shared a common tumor origin, supporting the heterogeneous scenario of acquired mechanisms of resistance (7). In addition, our group previous research found clonal evolution analyses suggest that the composition and relationship among resistant subclones, particularly relationship with T790M subclone, help to better understand the drug-resistant mechanism to TKIs (8).
Our case shows that combination therapy with icotinib and crizotinib can be safe and effective in patients with EGFR mutation and MET amplification detected by cfDNA as an acquired resistance mechanism. Also, the case highlights the reliable utility of noninvasive liquid biopsy assays into clinical practice to identify drivers of resistance to targeted therapies and the need for novel approaches to prevent and overcome the resistance mechanisms of targeted treatment. We are hopeful that an improved understanding of mechanisms of EGFR-TKIs resistance will lead to more treatment options for the emerging populations of EGFR-driven lung cancers.
Acknowledgments
We owe thanks to the patient and her family.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-442). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lettig L, Sahnane N, Pepe F, et al. EGFR T790M detection rate in lung adenocarcinomas at baseline using droplet digital PCR and validation by ultra-deep next generation sequencing. Transl Lung Cancer Res 2019;8:584-92. [Crossref] [PubMed]
- Campo M, Gerber D, Gainor JF, et al. Acquired Resistance to First-Line Afatinib and the Challenges of Prearranged Progression Biopsies. J Thorac Oncol 2016;11:2022-6. [Crossref] [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26. [Crossref] [PubMed]
- Baldacci S, Mazieres J, Tomasini P, et al. Outcome of EGFR-mutated NSCLC patients with MET-driven resistance to EGFR tyrosine kinase inhibitors. Oncotarget 2017;8:105103-14. [Crossref] [PubMed]
- Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. [Crossref] [PubMed]
- Chabon JJ, Simmons AD, Lovejoy AF, et al. Corrigendum: Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun 2016;7:13513. [Crossref] [PubMed]
- Chabon JJ, Simmons AD, Lovejoy AF, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nature communications 2016;7:11815. [Crossref] [PubMed]
- Jin Y, Bao H, Le X, et al. Distinct co-acquired alterations and genomic evolution during TKI treatment in non-small-cell lung cancer patients with or without acquired T790M mutation. Oncogene 2020;39:1846-59. [Crossref] [PubMed]