
PRRX1 isoform PRRX1A regulates the stemness phenotype and epithelial-mesenchymal transition (EMT) of cancer stem-like cells (CSCs) derived from non-small cell lung cancer (NSCLC)
Introduction
Lung cancer is the most common type of malignancy and one of the most fatal cancers globally. The 5-year survival rate of lung cancer is only approximately 20% (1,2). Non-small cell lung cancer (NSCLC) is the most common subtype of lung cancer, accounting for approximately 80–85% of all lung cancers. Although multiple strategies for curing NSCLC have been developed, including surgical resection, chemotherapy, and/or radiotherapy, unfortunately, most NSCLC patients suffer from chemoresistance and radioresistance, which leads to poor prognosis (3). At present, surgery remains the first choice for early stage disease, and platinum-based chemotherapy remains the standard procedure (3). Thus, efforts should be made to investigate the mechanisms inducing malignant behaviors in NSCLC.
Paired-related homeobox 1 (PRRX1), a transcription factor (TF), is tightly associated with tumorigenesis and malignancies (4-6). There are 2 alternative splicing transcripts of the PRRX1 gene: PRRX1A (245 amino acids) and PRRX1B (217 amino acids) (7,8). PRRX1A and PRRX1B are highly conserved and exert similar functions, including acting as TFs (9). In pancreatic regeneration and carcinogenesis processes, PRRX1A/B transcriptionally activates Sox9 and thus positively regulates Sox9-regulated acinar ductal metaplasia and regeneration. However, PRRX1A and PRRX1B function differently in these processes. For example, in pancreatic cancer, PRRX1A and PRRX1B fulfill separate functions and might synergize at distinct steps of physiological processes (10). We reasoned that there might be different functions of PRRX1A and PRRX1B in other cancers, since they are widely expressed.
PRRX1A and PRRX1B are involved in regulating the processes of epithelial-to-mesenchymal transition (EMT) and mesenchymal-to-epithelial transition (MET) (4,11), which modulate metastasis in liver cancers (10). This isoform-specific regulation of the EMT-MET axis has been revealed in breast (12), colorectal (13,14), and gastric cancer (15). The loss of PRRX1 was discovered to induce MET and metastatic colonization in breast cancer, potentially via loss of a PRRX1A-specific function or a decrease in the ratio of PRRX1A to PRRX1B (12), in the context of tumor progression. Therefore, the investigation into the potential functions and mechanisms of PRRX1A and PRRX1B and the identification of possible regulatory mechanisms will provide useful new insights into the biological roles of PRRX1A and PRRX1B.
Accumulating evidence has shown that tumors are composed of heterogeneous cell populations, including those capable of self-renewal and multilineage differentiation, defined as cancer stem-like cells (CSCs) (16), which contribute to the induction of drug-/radiation-resistance and recurrent metastases (17). In those CSCs derived from various kinds of cancer cells, CD24, CD44, and ALDH1 are considered specific markers (18-20). Several key stemness factors are critical for stemness maintenance in CSCs, including OCT4, Sox2, KLF4, and NANOG (21), which are tightly associated with malignancies. In adult neural stem/progenitor cells, SOX2 acts as a TF that is required for the maintenance of stemness in the central neuron system (22) and is coactivated with PRRX1 (23). Although there is insufficient evidence to demonstrate the exact mechanism of the coactivating roles between SOX2 and PRRX1, it is worth investigating whether PRRX1A and PRRX1B coactivate with SOX2 differently and thus affect stemness in CSCs derived from NSCLC.
In this study, we show that PRRX1A and PRRX1B present different expression patterns and exert different functions in regulating malignancies and stemness in CSCs derived from NSCLC. Bioinformatic analysis of clinical samples suggested that PRRX1A is correlated with the expression of TGF-β, which potentially causes EMT progression. Hence, in the current study, we explored whether PRRX1A affects the malignancy and stemness of CSCs derived from NSCLC.
We present the following article in accordance with the ARRVIE reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-633).
Methods
The experiments were approved by the Medical Ethics Committee of the First Affiliated Hospital of China Medical University. All animal experiments were performed in accordance with relevant guidelines of Animal Ethics Committee of the First Affiliated Hospital of China Medical University.
Cell culture and enrichment of CSCs
The human NSCLC A549 and SPC-A1 cell lines were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained in Dulbecco’s Modified Eagles Medium (DMEM, Gibco, Paisley, UK) supplemented with 10% fetal bovine serum (FBS), 100 units/mL of penicillin and 0.1 mg/mL of streptomycin (Gibco, Paisley, UK). Every 3 days, medium was refreshed or passaged.
For CSC culture, cells were maintained in DMEM/Ham Nutrient Mixture F-12 (1:1) with the addition of epidermal growth factor (EGF, 20 ng/mL), human fibroblast growth factor basic (hFGFb, 10 ng/mL), and 2% B27. Every 3 days, medium was half-replaced, and after 14–21 days, spheres were collected and stored in liquid nitrogen.
Tumor formation in nude mice
A total of 21 female BALB/c nude mice (4 weeks old) were kept in the specific-pathogen-free (SPF) facility. The mice were housed at room temperature with 12-h light/dark cycles, 50–65% humidity, and access to standard chow and water. All procedures in this section were approved by the Medical Ethics Committee of the Shanghai Outdo Biotech Company and performed according to the ethical guidelines. Briefly, mice were randomly divided into 3 groups, with 7 mice in each group. Next, 3×105 cells stably expressing pENTR/U6 vector, shSOX2, or shPRRX1A/B were injected under the back skin of the randomly grouped nude mice. From day 8 through day 28 after tumor inoculation, tumor size was measured for the longest (length) and the shortest (width) diameters every other day. Before either diameter reached 1 cm, mice were euthanized by inhalation of CO2 gas. The tumors were quickly dissected for imaging.
Serial replating assay
Cells were replated at clonal density (1,000 cells/well) and cultured in serum-free medium (SFM) supplemented with 2% B 27, 10 ng/mL EGF, and 20 ng/mL bFGF. Every 3 days, the medium was half-replaced. After 14 days, cells were then washed with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde in PBS, stained with 0.1% crystal violet (Sigma-Aldrich, St. Louis, MO, USA) for 10 min, washed with PBS, and the colonies were counted. For replating, same amount of cells were plated in SFM. After 14 days, the same procedure was performed and repeated 3 times.
Bioinformatics prediction
We used the Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn) to predict the levels of gene expression in NSCLC and normal tissues. It was also employed to access the correlation between genes.
cDNA microarray and quantitative real-time polymerase chain reaction (PCR)
A lung cancer cDNA microarray was purchased from Shanghai Outdo Biotech co., Ltd (Cat. No.: MecDNA-HLugA030PG01; Shanghai, China), which contained 30 paired cancer and paired adjacent lung tissues. The tissues were all from patients who were consecutive, newly diagnosed, untreated, and unselected (15 males, 15 females), and their ages ranged from 38 to 75 years (median age of 57 years). The experiments were approved by the Medical Ethics Committee of the Shanghai Outdo Biotech Company (ID: YBM-05-02). The cells were washed with ice-cold PBS twice and suspended with TRIZol reagent (Life Technologies, Grand Island, NY, USA), and total RNA was extracted by following the manufacturer’s instruction. Then, total RNA was dissolved in 40 µL of diethyl pyrocarbonate (DEPC) water and adjusted by 200 ng/mL at final concentration. For reverse transcription, 20 µL reaction system was prepared with a cDNA synthesis kit according to the manufacturer’s instruction. For quantitative polymerase chain reaction (qPCR), SYBR Green PCR Master Mix (Life Technologies, Grand Island, NY, USA) was used according to the manufacturer’s instruction. The involved primers were listed in Table 1.

Full table
The cDNA was used as template under the following conditions: 40 cycles of 95 °C for 30 s (s), 55 °C for 30 s, and 72 °C for 1 min. ABI7500 (Applied Biosystems, Foster City, CA, USA) was used for qPCR. Samples were compared using the relative CT method normalized to a housekeeping gene using 2−∆∆CT.
Western blot
Cells were suspended in Lysis buffer containing 50 mM Tris-HCl, 150 mM NaCl, 0.02% NaN3, 100 µg/mL Phenylmethanesulfonylfluoride fluoride (PMSF), 1 µg/mL aprotini, 1 µg/mL pepstatin A, and 1% Triton X-100. SoniConvert® homogenizer (DocSense, Chengdu, China) was employed to completely lyse cells. The following primary antibodies were used: Rabbit monoclonal anti-PRRX1 antibody (1:1,000, #ab211292); Rabbit monoclonal anti-TGF-β1 antibody (1:2,000, #ab92486); Rabbit monoclonal anti-E-cadherin antibody (1:1,000, #ab15148); Rabbit monoclonal anti-N-cadherin antibody (1:1,000, #ab18203); Rabbit monoclonal anti-vimentin antibody (1:1,000, #ab92547); Rabbit monoclonal anti-DDDK tag (binds to FLAG tag sequence) antibody (1:5,000, #ab1162); Rabbit monoclonal anti-SOX2 antibody (1:1,000, #ab93689); and Rabbit monoclonal anti-β-actin antibody (1:5,000, #ab8227). Goat anti-rabbit IgG H&L antibody (horseradish peroxidase-labeled, 1:10,000, #ab7090) was used as secondary antibody. Blot bands were quantified via densitometry with Image J software (National Institutes of Health Baltimore, MD, USA). β-actin was used as an internal reference.
Construction of expression plasmid and transfection
The full-length complementary DNA (cDNA) of PRRX1A (human PRRX1A, transcript variant pmx-1a; NCBI reference sequence: NM_006902.5) and PRRX1B (human PRRX1B, transcript variant pmx-1b; NCBI reference sequence: NM_022716.4) were obtained from RiboBio (Guangzhou, China) and ligated into the Hind III-Bam HI site of the p3×FLAG-CMV-10 vector (Sigma–Aldrich, St. Louis, MO, USA). The Hind III, Bam HI restriction enzymes, and T4 ligase were purchased from Takara (Heidelberg, Germany). Then, 0.8 µg of plasmid or empty vector was mixed with 4 µL Lipofectamine™ 2000 transfection reagent (Thermo Scientific, Waltham, MA, USA) in 0.5 mL OptiMEM medium (Thermo Scientific, Waltham, MA, USA), and this was applied to cells for 4 h followed by a medium-refresh. After 24 h, the culture medium was again refreshed.
Cell cycle analysis
To assess cell cycle distribution by quantification of DNA content, cells were harvested, washed with ice-cold PBS, and fixed overnight at 4 °C with ice-cold 70% ethanol. Then, fixed cells were washed with ice-cold PBS 3 times and incubated with a final concentration of 100 µg/mL RNase A and 40 µg/mL propidium iodide (PI, Beyotime) for 15 min in the dark. Then, cells were analyzed by 3 laser Navios flow cytometers (Beckman Coulter, Brea, CA, USA).
Transwell assay
Matrigel purchased from Millipore (Darmstadt, Germany) was diluted at a 1:3 concentration in SFM, and 50 µL of diluted Matrigel was added to the upper chamber of the Transwell plates and allowed to stand for 4 h. Single-cells were resuspended in SFM and seeded into the upper chamber at the amount of 1×104 cells/well. Subsequently, 600 µL of SFM supplemented with 20 ng/mL EGF, 10 ng/mL bFGF, and 2% B27 was added to the lower chamber. After 24-hour incubation, inserts were collected, and the cells on the lower face were fixed in 4% paraformaldehyde and stained with crystal violet for 15 min. The cells in 5 random views were counted.
Co-immunoprecipitation
For co-immunoprecipitation, 1×106 cells were lysed using radioimmunoprecipitation assay (RIPA) lysis buffer, and the protein concentration was measured by performing bicinchoninic acid (BCA) assay (Sigma) following the manufacturer’s instruction. Next, 50 µL of protein A agarose bead slurry was incubated with 10 µg of anti-DDDK tag antibody overnight at 4 °C with rotation. Then, the pellet beads were washed 3 times with PBS. The protein A/antibody complex was then incubated with 500 µg of lysate protein overnight with rotation, followed by 3 PBS washes. Then, the complex was heated at 100 °C for 10 mL, and 15 µL of eluted sample was loaded for the following semiquantitative Western blot assay.
Statistical analysis
All data are presented as mean ± SD (
Results
The expression pattern of CSCs derived from NSCLCs
After being cultured in SFM, epithelial A549 and SPC-A1 cells turned into spheres in suspension (Figure 1A). The proportion of CD133+ cells in parental A549 cells and SPC-A1 cells reached 7.2% or 5.3%, respectively, while the proportion of CD133+ cells in derived CSCs reached approximately 77.5% or 75.4% (Figure 1A). In the self-renewal assay, CSCs derived from both A549 and SPC-A1 cells showed continuous self-renewal capacity (Figure 1B). To determine the different expression patterns of EMT markers, PRRX1A, and PRRX1B among these cells, reverse transcriptase (RT)-qPCR assays were performed. It was found that compared to HBE cells, A549 cells presented no detectable differences in PRRX1A/B, EMT markers and, stemness factors, while SPC-A1 cells presented a lower expression of PRRX1B and EMT markers (Figure 1C). Remarkably, compared to parental cells, A549-CSCs and SPC-A1-CSCs had a significantly higher expression of PRRX1A and EMT markers (Figure 1D). The protein levels were consistent with their transcriptional levels (Figure 1E).
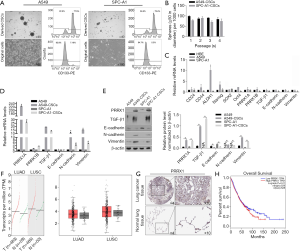
To evaluate the expression level of PRRX1 in pathological tissues, we performed an expression analysis by comparing lung adenocarcinoma (LUSC) and lung squamous cell carcinoma (LUSC) to normal tissues by using the GEPIA server. We found that PRRX1 was obviously but no significantly increased in NSCLC tissues (Figure 1F). Meanwhile, the human protein atlas verified an obvious expression trend of PRRX1 protein within NSCLC tissues (Figure 1G). Notably, PRRX1 expression was demonstrated to be irrelevant to prognosis (Figure 1H).
The potential roles of PRRX1A in regulating malignant behaviors in CSCs derived from NSCLC
To investigate the biological roles of PRRX1A, we overexpressed or downregulated PRRX1A or PRRX1B in A549-CSCs and SPC-A1-CSCs (Figure 2A,B), verified the efficiency, and observed changes in malignant behaviors. Propidium iodide (PI) staining followed by flow cytometry assays illustrated that, in both A549- and SPC-A1-CSCs, overexpression of PRRX1A but not PRRX1B promoted cell cycle progression from G1/G0 to S phase (Figure 2C). Notably, downregulation of both PRRX1A and PRRX1B blocked the cell cycle at G1/G0. By performing a CCK-8 assay, the promoting effect of overexpressed PRRX1A and the inhibitory effect of downregulated PRRX1A on cell viability were further confirmed (Figure 2D).
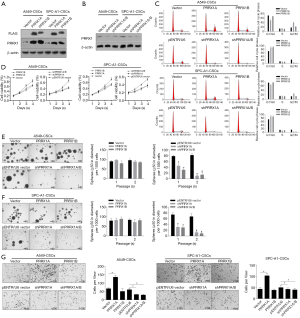
By considering the significantly higher expression levels of stemness factors and EMT markers in CSCs, we then determined whether PRRX1A/B expression was involved in regulating their processes. In the self-renewal capacity assay, knockdown of PRRX1A and PRRX1A/B obviously decreased self-renewal capacity in A549-CSCs and SPC-A1-CSCs (Figure 2E,F). Notably, both the overexpression of PRRX1A and PRRX1B failed to promote self-renewal capacity, potentially due to the endogenous protein level being sufficient. We also performed a Transwell invasion assay after overexpression or knockdown of PRRX1A/B. As illustrated in Figure 2G (left panel for A549-CSCs, right panel for SPC-A1-CSCs), knockdown of PRRX1A and PRRX1A/B significantly decreased the number of cells transferred to the other surface membrane. Surprisingly, overexpression of PRRX1A, but not PRRX1B, increased invasive capacity, indicating that PRRX1A, but not PRRX1B, plays a critical role in promoting invasiveness in both A549-CSCs and SPC-A1-CSCs.
Expression levels of PRRX1A and TGF-β in lung cancer cases
To determine the expression patterns of stemness factors, PRRX1A/B and EMT markers in lung cancer tissues compared to adjacent tissues, we examined their mRNA levels in 30 paired clinical lung cancer tissues (Figure 3A). We found that PRRX1A, PRRX1B, and TGF-β were significantly upregulated in lung cancer tissues compared to paired adjacent tissues, which indicated the potential correlation between all these genes. Furthermore, PRRX1A and TGF-β expression were positively correlated in lung cancer tissues, but there was no correlation with PRRX1B (Figure 3B).
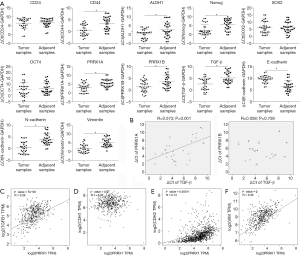
We also analyzed the correlation between PRRX1 and its target genes, including TGF-β1 by using the GEPIA server. Consistent with our clinical data, in 527 lung cancer clinical tissues, PRRX1 was positively and significantly correlated to TGF-β1 (Figure 3C). Despite of the irrelevance of E-cadherin with TGF-β1 (also termed CDH1, Figure 3D), TGF-β1 was also positively correlated with N-cadherin (also termed CDH2) and Vimentin (Figure 3E,F), which demonstrates that PRRX1 is tightly associated with TGF-β1 and EMT progression.
PRRX1A transcriptionally regulates TGF-β and subsequently modifies EMT processes
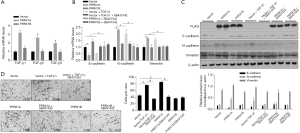
Transforming growth factor-β has 5 different isoforms, 3 of which are expressed in humans (TGF-β1, β2, and β3). This prompted us to assess the modulatory roles of overexpressed PRRX1A or PRRX1B in regulating all 3 isoforms. As presented in Figure 4A, the overexpression of PRRX1A significantly activated all 3 isoforms. To determine whether PRRX1A regulates EMT via the TGF-β signaling pathway, SB431542, a TGF-β receptor (TGF-βR) inhibitor, was employed to block TGF-β/TGF-βR signaling. By performing RT-qPCR (Figure 4B) and semiquantitative Western blotting (Figure 4C), it was observed that the regulation of EMT markers by overexpressed PRRX1A was reversed by the addition of SB43142, demonstrating that PRRX1A regulates EMT in a TGF-β/TGF-βR signaling pathway-dependent manner. Invasion assays further confirmed that PRRX1A promoted invasive capacity via the TGF-β/TGF-βR signaling pathway in both A549 CSCs and SPC-A1 CSCs (Figure 4D).
PRRX1A potentially regulates CSC stemness by stabilizing the stemness factor SOX2
In the RT-qPCR assay, A549-CSCs and SPC-A1-CSCs presented significantly higher levels of stemness factors than their parental cells (Figure 5A). To determine whether PRRX1A regulates CSC stemness, stemness factors were then measured. The results demonstrated that knockdown of PRRX1A and PRRX1A/B significantly decreased CD24, ALDH1, OCT4, Nanog, and Sox2 (Figure 5B, left panel). However, overexpression of PRRX1A or PRRX1B slightly affected stemness factors (Figure 5B, right panel), which was consistent with previous findings (Figure 2E,F). Shimozaki and colleagues reported that, in adult neural stem/progenitor cells, PRRX1A/B plays critical roles in maintaining stemness via binding to SOX2 on its HMG-box domain (23). This finding prompted us to confirm whether overexpressed PRRX1A and PRRX1B bind to SOX2 in A549 CSCs. As expected, overexpressed PRRX1A strongly bound to the SOX2 protein (Figure 5C), and overexpressed PRRX1B was also observed to bind to PRRX1B, but it is unknown whether PRRX1B binds directly or indirectly to SOX2. SOX2 protein detection revealed that PRRX1A overexpression increased SOX2 protein by potentially stabilizing SOX2, which was not affected by the blocking of the TGF-β/TGF-βR signaling pathway (Figure 5D).
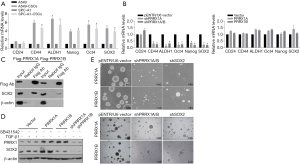
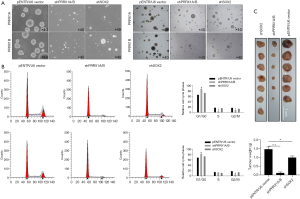
Sphere formation assays revealed that knockdown of both PRRX1A/B and SOX2 obviously decreased sphere formation ability in both A549-CSCs and SPC-A1-CSCs (Figure 6A). To further confirm whether PRRX1A and SOX2 maintained stemness via regulating cell proliferation, the proportion of cells in each cell cycle phase was assessed. Knockdown of PRRX1A, but not SOX2 affected cell cycle distribution significantly, indicating that PRRX1A contributes to stemness maintenance partially via the stabilizing of SOX2 (Figure 6B). To gain additional evidence in vivo, tumor formation in nude mice was analyzed. By injecting stable transfected CSCs into the backs of nude mice, we observed that both PRRX1A/B and SOX2 knockdown significantly decreased the tumor size and weight when compared to the control group (Figure 6C).
Discussion
We have shown that one of the isoforms of PRRX1, PRRX1A, but not PRRX1B, enhances the mesenchymal phenotype of CSCs derived from NSCLC via transcriptionally activating TGF-β, revealing a dependence on the TGF-β/TGF-βR signaling pathway. Knockdown of endogenous PRRX1A and PRRX1A/B obviously decreased the stemness of CSCs, which was further confirmed to exert a regulatory role by interacting with SOX2 and thus stabilizing it. This result suggests that PRRX1A, at least partially, is critical for maintaining stemness. Our data on CSCs agree with several reports that showed that PRRX1 induces EMT progression and thus regulates metastasis and results in poor prognosis (13) and that PRRX1 contributes to maintaining stemness in CSCs derived from breast cancer (5). However, in all these reports, whether the 2 isoforms of PRRX1, PRRX1A and PRRX1B, contribute differently was not further investigated. In the current study, PRRX1A was found to play more critical roles in regulating EMT and stemness of CSCs. Higher expression levels of PRRX1A than PRRX1B in clinical NSCLC tissues also indicated that PRRX1A may play a more critical role in carcinogenesis and tumor progression. The roles of PRRX1B in regulating EMT and stemness properties might be diverse, and further studies are required.
In a previous report, PRRX1A was identified as a transcriptional activator, and PRRX1B was found to exert the opposing function as a transcriptional repressor (24). Consistently, in our results, overexpression of PRRX1A transcriptionally activated the TGF-β family and promoted EMT progression in a TGF-β/TGF-βR signaling pathway-dependent manner. However, overexpression of PRRX1B failed to obviously affect TGF-β expression. This observation suggests that endogenous PRRX1B may be irrelevant to the regulation of the TGF-β family in the context of CSC biology. One possibility is that the functional characteristics of PRRX1A and PRRX1B are cell type-specific. Due to the difficulty of specifically decreasing PRRX1B without disturbing PRRX1A, we failed to provide supporting evidence showing whether PRRX1B is relevant to TGF-β activation. This is worth investigating in further research.
Increasing evidence supports that the balance of EMT-MET progression regulates cancer metastasis (25). Here, downregulation of epithelial E-cadherin and upregulation of mesenchymal N-cadherin and Vimentin were apparent when PRRX1A, but not PRRX1B, was overexpressed, while a consistent invasive phenotype was observed. PRRX1A exhibited precipitative effects in EMT, which is consistent with a recent report revealing the critical roles of PRRX1 in EMT progression (5). In that report, Shi and colleagues reported that PRRX1 regulates the balance of EMT-MET via regulating KLF4, a stemness factor, and subsequently causes malignancy and stemness (5). In this study, a coimmunoprecipitation assay showed that PRRX1A and PRRX1B interact with the SOX2 protein. Compared to PRRX1B, PRRX1A presents an obviously stronger interaction with SOX2, indicating that PRRX1B might form a heterotrimer with PRRX1A and SOX2 instead of interacting directly with SOX2. The binding of PRRX1A to SOX2 stabilized SOX2 and promoted stemness in CSCs, while knockdown of SOX2 or PRRX1A obviously decreased stemness in CSCs, indicating that PRRX1A potentially regulates stemness in CSCs via regulating SOX2. Thus, our results revealed a novel mechanism by which PRRX1 regulates the stemness of CSCs. We failed to uncover the specific roles of PRRX1B, which may be due to our focus on CSC biology.
Our findings collectively show that PRRX1A exerts dual functions via different mechanisms; however, PRRX1B was not found to be involved in these processes. Targeting the PRRX1A-TGF-β-TGF-βR and PRRX1A-SOX2 axes may offer a new therapeutic approach in curing metastatic NSCLC. These results not only shed light on the role of PRRX1A in regulating CSCs derived from NSCLC but also further reveal the different roles of PRRX1A and PRRX1B in CSC origination, malignant transformation, and metastasis.
Acknowledgments
We would like to thank Ziyi Zhao, Qiongying Hu, and Changjin Chen for their technical support.
Funding: This work was financially supported by the Program Funded by Liaoning Province Education Administration (no. QN2019003).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-63
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-63
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-63). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The experiments were approved by the Medical Ethics Committee of the First Affiliated Hospital of China Medical University. All animal experiments were performed in accordance with relevant guidelines of Animal Ethics Committee of the First Affiliated Hospital of China Medical University.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet 2015;385:977-1010. [Crossref] [PubMed]
- Zaman A, Bivona TG. Emerging application of genomics-guided therapeutics in personalized lung cancer treatment. Ann Transl Med 2018;6:160. [Crossref] [PubMed]
- Gregg JP, Li T, Yoneda KY. Molecular testing strategies in non-small cell lung cancer: optimizing the diagnostic journey. Transl Lung Cancer Res 2019;8:286-301. [Crossref] [PubMed]
- Marchand B, Pitarresi JR, Reichert M, et al. PRRX1 isoforms cooperate with FOXM1 to regulate the DNA damage response in pancreatic cancer cells. Oncogene 2019;38:4325-39. [Crossref] [PubMed]
- Shi L, Tang X, Qian M, et al. A SIRT1-centered circuitry regulates breast cancer stemness and metastasis. Oncogene 2018;37:6299-315. [Crossref] [PubMed]
- Chen X, Wu L, Li D, et al. Radiosensitizing effects of miR-18a-5p on lung cancer stem-like cells via downregulating both ATM and HIF-1α. Cancer Med 2018;7:3834-47. [Crossref] [PubMed]
- Kern MJ, Witte DP, Valerius MT, et al. A novel murine homeobox gene isolated by a tissue specific PCR cloning strategy. Nucleic Acids Res 1992;20:5189-95. [Crossref] [PubMed]
- Kern MJ, Argao EA, Birkenmeier EH, et al. Genomic organization and chromosome localization of the murine homeobox gene Pmx. Genomics 1994;19:334-40. [Crossref] [PubMed]
- Reichert M, Takano S, von Burstin J, et al. The Prrx1 homeodomain transcription factor plays a central role in pancreatic regeneration and carcinogenesis. Genes Dev 2013;27:288-300. [Crossref] [PubMed]
- Takano S, Reichert M, Bakir B, et al. Prrx1 isoform switching regulates pancreatic cancer invasion and metastatic colonization. Genes Dev 2016;30:233-47. [Crossref] [PubMed]
- Ocaña OH, Coskun H, Minguillón C, et al. A right-handed signalling pathway drives heart looping in vertebrates. Nature 2017;549:86-90. [Crossref] [PubMed]
- Ocaña OH, Córcoles R, Fabra A, et al. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell 2012;22:709-24. [Crossref] [PubMed]
- Takahashi Y, Sawada G, Kurashige J, et al. Paired related homoeobox 1, a new EMT inducer, is involved in metastasis and poor prognosis in colorectal cancer. Br J Cancer 2013;109:307-11. [Crossref] [PubMed]
- Zheng L, Zhang Y, Lin S, et al. Down-regualtion of miR-106b induces epithelial-mesenchymal transition but suppresses metastatic colonization by targeting Prrx1 in colorectal cancer. Int J Clin Exp Pathol 2015;8:10534-44. [PubMed]
- Guo J, Fu Z, Wei J, et al. PRRX1 promotes epithelial-mesenchymal transition through the Wnt/β-catenin pathway in gastric cancer. Med Oncol 2015;32:393. [Crossref] [PubMed]
- Mao H. Clinical relevance of mutant-allele tumor heterogeneity and lung adenocarcinoma. Ann Transl Med 2019;7:432. [Crossref] [PubMed]
- Ellsworth RE, Blackburn HL, Shriver CD, et al. Molecular heterogeneity in breast cancer: State of the science and implications for patient care. Semin Cell Dev Biol 2017;64:65-72. [Crossref] [PubMed]
- Valeta-Magara A, Gadi A, Volta V, et al. Inflammatory Breast Cancer Promotes Development of M2 Tumor-Associated Macrophages and Cancer Mesenchymal Cells through a Complex Chemokine Network. Cancer Res 2019;79:3360-71. [Crossref] [PubMed]
- Hu J, Guan W, Yan L, et al. Cancer Stem Cell Marker Endoglin (CD105) Induces Epithelial Mesenchymal Transition (EMT) but Not Metastasis in Clear Cell Renal Cell Carcinoma. Stem Cells Int 2019;2019:9060152. [Crossref] [PubMed]
- Son MJ, Woolard K, Nam DH, et al. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell 2009;4:440-52. [Crossref] [PubMed]
- Finicelli M, Benedetti G, Squillaro T, et al. Expression of stemness genes in primary breast cancer tissues: the role of SOX2 as a prognostic marker for detection of early recurrence. Oncotarget 2014;5:9678-88. [Crossref] [PubMed]
- Ferri AL, Cavallaro M, Braida D, et al. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development 2004;131:3805-19. [Crossref] [PubMed]
- Shimozaki K, Clemenson GD Jr, Gage FH. Paired related homeobox protein 1 is a regulator of stemness in adult neural stem/progenitor cells. J Neurosci 2013;33:4066-75. [Crossref] [PubMed]
- Norris RA, Kern MJ. The identification of Prx1 transcription regulatory domains provides a mechanism for unequal compensation by the Prx1 and Prx2 loci. J Biol Chem 2001;276:26829-37. [Crossref] [PubMed]
- Jolly MK, Boareto M, Huang B, et al. Implications of the Hybrid Epithelial/Mesenchymal Phenotype in Metastasis. Front Oncol 2015;5:155. [Crossref] [PubMed]


