
Simultaneous targeting of MET overexpression in EGFR mutation-positive non-small cell lung cancer can increase the benefit of EGFR-TKI therapy?
The hepatocyte growth factor receptor (MET) is a receptor tyrosine kinase that is activated by binding of its ligand, hepatocyte growth factor (HGF), and which triggers signaling via the RAS-MEK-ERK, PI3K-AKT, Wnt-β-catenin, and STAT pathways (1). The extracellular region of MET contains semaphorin, cysteine-rich, and immunoglobulin domains, and the intracellular region comprises a juxtamembrane domain, the tyrosine kinase catalytic domain, and a carboxyl-terminal docking site (1). MET is a proto-oncogene, and dysregulation of MET signaling in lung cancer occurs through a variety of mechanisms, including gene mutation, amplification, and rearrangement as well as protein overexpression (1). MET amplification (METamp) is thought to increase MET signaling as a result of the associated protein overexpression and constitutive kinase activation. De novo METamp has been detected in ~1% to 5% of lung adenocarcinomas and ~1% of squamous cell lung cancers (1-3). Individuals with non–small cell lung cancer (NSCLC) positive for activating mutations of the epidermal growth factor receptor gene (EGFR) receive clinical benefit from treatment with EGFR tyrosine kinase inhibitors (TKIs) (4). However, such patients eventually develop resistance to these drugs, with the mechanism of acquired resistance being the development of a secondary T790M mutation of EGFR in ~60% of cases (4). METamp has also been identified as a mechanism of acquired resistance to first-, second-, and third-generation EGFR-TKIs in patients with EGFR-mutated NSCLC (4). Conversely, preclinical studies have shown that MET-amplified lung cancer cells exposed to MET inhibitors for a prolonged period develop resistance to these agents through up-regulation of the EGFR signaling pathway (5). Given this background, Scagliotti and colleagues hypothesized that the addition of a MET inhibitor to an EGFR-TKI might prolong progression-free survival (PFS) in EGFR-mutated NSCLC by delaying treatment-emergent EGFR-TKI resistance due to MET signaling (6).
These researchers thus designed a randomized, controlled phase 2 study to evaluate the potential benefit of combination treatment with the MET inhibitor emibetuzumab and the first-generation EGFR-TKI erlotinib in chemotherapy-naïve patients with EGFR mutation–positive NSCLC. No significant difference in median PFS was detected between patients receiving both drugs and those receiving erlotinib alone in the intention-to-treat population, and the study did not meet its primary end point. However, exploratory analysis based on MET expression in tumor cells revealed that patients with a high level of MET expression (MET immunohistochemistry score of 3+ in at least 90% of tumor cells) might receive a clinically meaningful PFS benefit from the addition of emibetuzumab to erlotinib (median PFS of 20.7 versus 5.4 months). Given that an analysis of baseline characteristics in this patient subpopulation did not show any imbalance between treatment arms with regard to covariates known to be of prognostic relevance in EGFR-mutant NSCLC patients, and that the MET-high patients showed a substantially shorter median PFS during erlotinib treatment compared with the corresponding MET-low patients, the findings of this study indeed suggest that there is potential benefit of adding emibetuzumab to erlotinib for EGFR mutation–positive NSCLC with a high level of MET expression. However, the results must be carefully interpreted according to the level of MET expression. Exploratory post-hoc analysis showed that the PFS improvement was relevant in only 12 of 71 patients (17%) with the highest MET expression level (MET score of 3+ in ≥90% of tumor cells). It will be necessary to confirm that staining intensity and the cutoff value are reproducible and can be standardized.
MET status in clinical trials has been defined mainly by three tests: immunohistochemistry (IHC) for detection of MET protein overexpression, fluorescence in situ hybridization (FISH) for detection of MET copy number alterations (CNAs) including METamp, and next-generation sequencing (NGS) analysis of MET mutations including exon-14 (METex14) alterations. The frequency of MET protein overexpression in NSCLC is variable, ranging from 5% to 75% (7), and the finding by Tsuta et al. that ~60% of their patients had a MET IHC score of ≥2+ in ≥60% of tumor cells is compatible with previous reports. MET IHC has led to conflicting results regarding the role of MET as a predictive biomarker in several previous trials, given that MET protein overexpression does not always reflect increased MET receptor activation (8). In addition, the frequency of dual positivity for MET overexpression and MET CNA in NSCLC specimens was found to be only ~30% (8). Indeed, MET IHC appears to be an inefficient screen for METamp or for METex14 alterations (9).
Although FISH analysis has been performed to investigate MET CNA in NSCLC, there is no consensus on the definition of MET CNA (3,10). The definition has thus been based on the number of MET signals per cell [MET gene copy number (GCN), Cappuzzo scoring system] or on the ratio of the copy number for MET to that of chromosome 7 (MET/CEP7 ratio) (3). METamp is defined by MET GCN or the MET/CEP7 ratio. About 20% of NSCLC patients with METex14 alterations were found to be positive for concurrent high-level METamp (MET/CEP7 ratio of ≥3) in surgically resected tumor specimens, and these genomic alterations were associated with a poorer prognosis (10,11). Patients with lung adenocarcinoma positive for high-level METamp (MET/CEP7 ratio of ≥5) were found not to harbor concurrent driver mutations in known oncogenes (EGFR, KRAS, ALK, ERBB2, BRAF, NRAS, ROS1, or RET) (12). A high MET CNA represents the best case for a true MET copy number gain-dependent MET-driven state.
MET IHC depends on the pathologist performing the analysis and is not readily standardized. The MET expression cutoffs based on increments of 10% of positive tumor cells adopted in the study by Scagliotti and colleagues are thus likely not to be highly reproducible. In a phase Ib/II study of combined treatment with the MET inhibitor capmatinib and the first-generation EGFR-TKI gefitinib after failure of EGFR-TKI monotherapy in patients with EGFR-mutated and MET-dysregulated NSCLC, MET GCN was selected as a biomarker because the response correlated better with MET GCN (with a cutoff of ≥6) than with the MET IHC score (13).
The promising data of the INSIGHT (14,15) and TATTON (16) studies is expected to spur the further pursuit of treatment with a MET inhibitor in combination with an EGFR-TKI in patients with EGFR-mutated advanced NSCLC positive for METamp after the development of EGFR-TKI resistance. The third-generation EGFR-TKI osimertinib has recently become established as a new standard of care in the first-line setting for patients with NSCLC harboring EGFR mutations, on the basis of a pivotal phase III trial (FLAURA trial) showing that osimertinib monotherapy conferred a significantly longer PFS compared with the first-generation EGFR-TKIs gefitinib or erlotinib (17). METamp was the most common mechanism underlying acquired resistance to first-line osimertinib, being detected in ~15% of patients by NGS of circulating DNA (4,18). Given this background, several clinical trials (including SAVANNAH and ORCHARD) designed to assess the combination of a MET inhibitor and osimertinib after the development of METamp-mediated resistance to osimertinib are underway. There are currently no approved targeted therapies for NSCLC positive for METamp (Table 1).
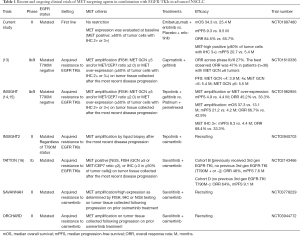
Full table
In contrast to treatment for METamp, molecularly targeted therapy for lung adenocarcinoma harboring a METex14 skipping mutation has been introduced into clinical practice. METex14 alterations were initially identified in SCLC and NSCLC in 2003 and 2005, respectively (19). METex14 encodes the juxtamembrane domain and tyrosine-1003 residue that serves as the binding site for CBL, an E3 ubiquitin ligase that controls MET turnover. Ubiquitination of MET thus results in its internalization and degradation and thereby attenuates its promotion of cell survival and proliferation. METex14 mutations that disrupt splice sites flanking the exon result in aberrant splicing and exon skipping. The resulting mutant protein is less susceptible to ubiquitination and consequent degradation, resulting in sustained MET activation and oncogenesis (1,2). METex14 alterations have been detected in 4.3% of lung adenocarcinomas and in 3.0% of squamous cell lung cancers (2). Lung adenocarcinomas harboring METex14 alterations manifest a substantial clinical response to MET inhibition (2,20). These mutations thus join those in EGFR and ALK as targetable driver alterations that occur in a not insignificant proportion of lung cancer patients (8). Capmatinib was approved by the U.S. Food and Drug Administration in May 2020 for the treatment of advanced NSCLC positive for METex14 skipping mutations on the basis of the GEOMETRY mono-1 phase II trial (21) (Table 2). The MET inhibitor tepotinib was similarly approved in Japan in March 2020 on the basis of the results of the VISION phase II trial (24).
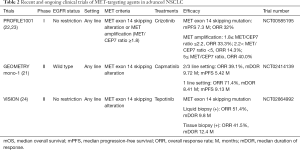
Full table
There are several limitations to the study of Scagliotti et al. First, osimertinib has supplanted gefitinib and erlotinib for first-line treatment of patients with NSCLC harboring EGFR mutations (14). However, the same strategy may be applicable to patients treated with osimertinib. Although there are no data with regard to how the MET pathway might be affected by osimertinib treatment, the concept of adding emibetuzumab to osimertinib in the same setting thus warrants further investigation. A second limitation of the study relates to MET biomarker selection. Accurate biomarker selection is necessary to identify patients who are expected to benefit from emibetuzumab. Although MET IHC was selected as the biomarker in this trial, this method is more difficult to standardize with clear criteria than is FISH analysis of MET CNA including METamp. It might actually be necessary to combine several test methods for determination of MET status so as not to overlook patients with MET dysregulation. In the TATTON trial, three test methods—IHC, FISH, and NGS—were adopted to detect MET dysregulation, and the results of the three tests did not overlap completely (16). MET IHC alone thus cannot be considered a reliable biomarker for prediction of emibetuzumab efficacy.
In conclusion, the study by Scagliotti and colleagues showed that the combination of emibetuzumab and erlotinib provided a clinically meaningful benefit in first-line treatment of the subgroup of EGFR-mutated NSCLC patients whose tumors express MET at a high level. The translation of this finding to actual clinical practice will require establishment of an optimal predictive biomarker for MET-targeted therapy.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-707). MT reports honoraria from Boehringer Ingelheim, Chugai Pharmaceutical, AstraZeneca, Ono Pharmaceutical, and Novartis during the conduct of the study. MT serves as an unpaid editorial board member of Translational Lung Cancer Research from August 2019 to August 2021. KN reports grants and personal fees from AstraZeneca K.K., grants and personal fees from Astellas Pharma Inc., grants and personal fees from MSD K.K., grants, personal fees and other from Ono Pharmaceutical Co., Ltd., grants and personal fees from Nippon Boehringer Ingelheim Co., Ltd., grants and personal fees from Novartis Pharma K.K., grants, personal fees and other from Pfizer Japan Inc., grants and personal fees from Bristol Myers Squibb Company, grants, personal fees and other from Eli Lilly Japan K.K., grants and personal fees from Chugai Pharmaceutical Co., Ltd., grants and personal fees from Daiichi Sankyo Co., Ltd., grants and personal fees from Merck Serono Co., Ltd.,/ Merck Biopharma Co., Ltd., during the conduct of the study; personal fees from Clinical Trial Co., Ltd., personal fees from MEDICUS SHUPPAN, Publishers Co., Ltd., personal fees from Care Net, Inc., personal fees from Reno. Medical K.K., personal fees and other from KYORIN Pharmaceutical Co., Ltd., personal fees from Medical Review Co., Ltd., personal fees from Roche Diagnostics K.K., personal fees from Bayer Yakuhin, Ltd., personal fees from Medical Mobile Communications co., Ltd., personal fees from 3H Clinical Trial Inc., personal fees from Nichi-Iko Pharmaceutical Co., Ltd., grants, personal fees and other from Takeda Pharmaceutical Co., Ltd., grants and personal fees from Taiho Pharmaceutical Co., Ltd., grants and personal fees from SymBio Pharmaceuticals Limited., personal fees from NANZANDO Co., Ltd., personal fees from YODOSHA CO., LTD., personal fees from Nikkei Business Publications, Inc., personal fees from Thermo Fisher Scientific K.K., personal fees from YOMIURI TELECASTING CORPORATION., personal fees from Nippon Kayaku Co., Ltd., grants and personal fees from AbbVie Inc, grants from inVentiv Health Japan, grants from ICON Japan K.K., grants from GRITSONE ONCOLOGY.INC, grants from PAREXEL International Corp., grants from Kissei Pharmaceutical Co., Ltd., grants from EPS Corporation., grants from Syneos Health., grants from Pfizer R&D Japan G.K., grants from A2 Healthcare Corp., grants from Quintiles Inc./IQVIA Services JAPAN K.K., grants from EP-CRSU CO., LTD., grants from Linical Co., Ltd., grants from Eisai Co., Ltd., grants from CMIC Shift Zero K.K., grants from Kyowa Hakko Kirin Co., Ltd., grants from Bayer Yakuhin, Ltd, grants from EPS International Co., Ltd., grants from Otsuka Pharmaceutical Co., Ltd., outside the submitted work. HK has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Drilon A, Cappuzzo F, Ou SH, et al. Targeting MET in Lung Cancer: Will Expectations Finally Be MET? J Thorac Oncol 2017;12:15-26. [Crossref] [PubMed]
- Paik PK, Drilon A, Fan PD, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring met mutations causing exon 14 skipping. Cancer Discov 2015;5:842-9. [Crossref] [PubMed]
- Cappuzzo F, Marchetti A, Skokan M, et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol 2009;27:1667-74. [Crossref] [PubMed]
- Westover D, Zugazagoitia J, Cho BC, et al. Mechanisms of acquired resistance to first-and second-generation EGFR tyrosine kinase inhibitors. Ann Oncol 2018;29:i10-9. [Crossref] [PubMed]
- McDermott U, Pusapati R V., Christensen JG, et al. Acquired resistance of non-small cell lung cancer cells to MET kinase inhibition is mediated by a switch to epidermal growth factor receptor dependency. Cancer Res 2010;70:1625-34. [Crossref] [PubMed]
- Scagliotti G, Moro-Sibilot D, Kollmeier J, et al. A Randomized-Controlled Phase 2 Study of the MET Antibody Emibetuzumab in Combination with Erlotinib as First-Line Treatment for EGFR Mutation–Positive NSCLC Patients. J Thorac Oncol 2020;15:80-90. [Crossref] [PubMed]
- Park S, Choi Y, La , Sung CO, et al. High MET copy number and MET overexpression: Poor outcome in non-small cell lung cancer patients. Histol Histopathol 2012;27:197-207. [PubMed]
- Tsuta K, Kozu Y, Mimae T, et al. C-MET/phospho-MET protein expression and MET gene copy number in non-small cell lung carcinomas. J Thorac Oncol 2012;7:331-9. [Crossref] [PubMed]
- Guo R, Berry LD, Aisner DL, et al. MET IHC Is a Poor Screen for MET Amplification or MET Exon 14 Mutations in Lung Adenocarcinomas : Data from a Tri-Institutional Cohort of the Lung Cancer Mutation Consortium. J Thorac Oncol 2019;14:1666-71. [Crossref] [PubMed]
- Tong JH, Yeung SF, Chan AWH, et al. MET amplification and exon 14 splice site mutation define unique molecular subgroups of non-small cell lung carcinoma with poor prognosis. Clin Cancer Res 2016;22:3048-56. [Crossref] [PubMed]
- Awad MM, Oxnard GR, Jackman DM, et al. MET exon 14 mutations in Non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-Met overexpression. J Clin Oncol 2016;34:721-30. [Crossref] [PubMed]
- Noonan SA, Berry L, Lu X, et al. Identifying the appropriate FISH criteria for defining MET copy number-driven lung adenocarcinoma through oncogene overlap analysis. J Thorac Oncol 2016;11:1293-304. [Crossref] [PubMed]
- Wu YL, Zhang L, Kim DW, et al. Phase Ib/II Study of Capmatinib (INC280) Plus Gefitinib after Failure of Epidermal Growth Factor Receptor (EGFR) Inhibitor Therapy in Patients with EGFR-Mutated, MET Factor-Dysregulated Non-Small-Cell Lung Cancer. J Clin Oncol 2018;36:3101-9. [Crossref] [PubMed]
- Park K, Zhou J, Kim D-W, et al. Tepotinib plus gefitinib in patients with MET-amplified EGFR-mutant NSCLC: Long-term outcomes of the INSIGHT study. Ann Oncol 2019;30:ix159. [Crossref]
- Cheng Y, Zhou J, Lu S, et al. Phase II study of tepotinib + gefitinib (TEP+GEF) in MET-positive (MET+)/epidermal growth factor receptor (EGFR)-mutant (MT) non-small cell lung cancer (NSCLC). Ann Oncol 2018;29:viii493-viii547.
- Sequist LV, Han JY, Ahn MJ, et al. Osimertinib plus savolitinib in patients with EGFR mutation-positive, MET-amplified, non-small-cell lung cancer after progression on EGFR tyrosine kinase inhibitors: interim results from a multicentre, open-label, phase 1b study. Lancet Oncol 2020;21:373-86. [Crossref] [PubMed]
- Ramalingam SS, Vansteenkiste J, Planchard D, et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N Engl J Med 2020;382:41-50. [Crossref] [PubMed]
- Cho BC, Cheng Y, Zhou C, et al. Mechanisms of acquired resistance to first-line osimertinib: Preliminary data from the phase III FLAURA study. Ann Oncol 2018;29:ix177. [Crossref]
- Comoglio PM, Trusolino L, Boccaccio C. Known and novel roles of the MET oncogene in cancer: A coherent approach to targeted therapy. Nat Rev Cancer 2018;18:341-58. [Crossref] [PubMed]
- Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov 2015;5:850-9. [Crossref] [PubMed]
- Wolf J, Seto T, Han J-Y, et al. Capmatinib (INC280) in METΔex14-mutated advanced non-small cell lung cancer (NSCLC): Efficacy data from the phase II GEOMETRY mono-1 study. J Clin Oncol 2019;37:abstr 9004.
- Drilon A, Clark J, Weiss J, et al. OA12.02 Updated Antitumor Activity of Crizotinib in Patients with MET Exon 14-Altered Advanced Non-Small Cell Lung Cancer. J Thorac Oncol 2018;13:S348. [Crossref]
- Camidge DR, Otterson GA, Clark JW, et al. Crizotinib in patients (pts) with MET-amplified non-small cell lung cancer (NSCLC): Updated safety and efficacy findings from a phase 1 trial. J Clin Oncol 2018;36:abstr 9062.
- Paik PK, Veillon R, Cortot AB, et al. Phase II study of tepotinib in NSCLC patients with METex14 mutations. J Clin Oncol 2019;37:abstr 9005.


