
Concomitant genetic alterations are associated with response to EGFR targeted therapy in patients with lung adenocarcinoma
Introduction
For patients with advanced lung adenocarcinoma (LADC), the discovery of epidermal growth factor receptor (EGFR) mutations and their sensitivity to tyrosine kinase inhibitors (TKIs) changed the landscape of treatment regimens (1-3). So far, gefitinib, erlotinib and afatinib are still the best choice for first-line treatment of patients with EGFR mutations in non-small cell lung cancer (NSCLC) (4) However, acquired resistance usually occurs 10–12 months after treatment with EGFR-TKIss. Third-generation EGFR-TKIs can specifically target EGFR-T790M and EGFR activation mutations and show significant activity in EGFR-T790M positive NSCLC patients with acquired drug resistance (5,6). Unfortunately, resistance to the third-generation EGFR-TKIs is also inevitable. Some studies have demonstrated that by quantifying EGFR mutation alleles the response to EGFR-TKIs can be predicted (7,8), and others have shown that different EGFR mutations can affect the prognosis of patients who receive EGFR-TKI treatment (9,10). However, this approach does not address the potential risk of co-occurring genetic alterations in cancer, but rather excludes a “driving factor” from other factors. Existing studies have suggested that the current single-gene driver-oncogene viewpoint needs to be reviewed, and clinical outcomes should be related to accompanying genetic changes in patients with advanced lung cancer such as EGFR mutation (11-13). At present, the prevalence of co-occurring genetic changes affecting clinical outcomes in advanced lung cancer with major carcinogenic drivers is largely unknown. Therefore, to optimize clinical outcomes of EGFR-TKI therapy, a better understanding of the underlying mechanism of this phenomenon is needed.
Dynamic monitoring of tumor genomic profiles is of great importance to the diagnosis, treatment, and prognosis of patients with NSCLC (14,15). To better identify these underlying mechanisms, we performed mutation profiling analysis on 36 plasma samples of NSCLC lung adenocarcinoma before further treatment, and targeted 556 cancer-related genes using next-generation sequencing.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-679).
Methods
Patient selection
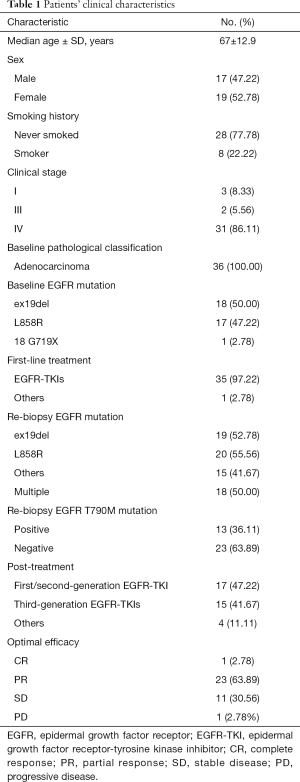
Thirty-six patients who were initially diagnosed with lung adenocarcinoma at the Affiliated Hospital of Guangdong Medical University between May, 2017 and December, 2019 were enrolled. And their medical records were retrospectively reviewed. Table 1 shows a summary of the patients’ clinical characteristics.
Full table
The inclusion criteria were as follows: (I) 18–80 years old; (II) initial diagnosis of lung adenocarcinoma, and EGFR mutation confirmed by molecular pathology, detected using an amplification refractory mutation system (ARMS); (III) clinical acceptance of first and second-generation EGFR-TKI treatment; (IV) clinicians believed re-biopsy necessary to guide the treatment based on the response evaluation criteria in solid tumors (RECIST); and (V) an Eastern Cooperative Oncology Group (ECOG), performance status (PS) score of ≤2.
This study was approved by the Ethics Committee of the Affiliated Hospital of Guangdong Medical University. Informed consent was obtained from all patients who underwent blood genetic analysis. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013).
Sample preparation, targeted NGS, and data processing
The liquid biopsy specimens were collected into two catheters using standard venipuncture techniques. Circulating tumor DNA (ctDNA) was extracted from EDTA blood specimen plasma by following the recommended method (QIAamp Circulating Nucleic Acid Kit; Qiagen, Germantown, MD, USA), and quantified by Qubit 4.0 using the dsDNA HS Assay Kit (ThermoFisher Scientific). DNA shearing is conducted per protocol and a quality control check is performed. Average fragment size should be 150–250 bp. Sequence libraries were prepared using the Ion AmpliSeq Library Kit (Thermo Fisher, Waltham, MA, USA). The blunt 5'terminal phosphorylation fragment was produced first. The dAMP was added (A-tailing) to the 3' ends of the dsDNA library fragments. Next, ligate the dsDNA adapter with 3'dtmp to the a-tailed library fragment Library fragments with appropriate adapter sequences are amplified via ligation-mediated pre-capture PCR. The libraries were then pooled and sequenced using the NovaSeq 6000 Sequencing Instruments (Illumina Inc., Austin, TX, USA) and reagents as well as PhiX Control v3 (Illumina). Sequencing data were mapped using Burrows-Wheeler Aligner (BWA-MEM, v0.7.12) (16). Local realignment was performed around indels and base quality score to recalibrate using the Genomic Analysis Toolkit (GATK 3.4.0) (Broad Institute, Cambridge, MA, USA) (17). MuTect 2 (Broad Institute, Cambridge, MA, USA) and Vardict were employed to detect somatic mutations (18).
Follow-up
Progression-free survival (PFS) was defined as the duration from the initiation of EGFR-TKI treatment to disease progression or death from any cause. Clinical follow-up assessments took place every four weeks. Tumor response was evaluated by experienced researchers based on the RECIST guidelines (version 1.1), and was divided into complete response (CR), partial response (PR), stable disease (SD), or progressive disease (PD). The cut-off date for analysis was February, 25, 2020.
Statistical analysis
All statistical analyses were performed using SPSS v25 (IBM Corporation, NY, USA). The Kaplan-Meier method was employed to estimate survival, and differences between variables were compared using the log-rank test. P values were calculated using Fisher’s exact test and Pearson’s test for categorical and continuous variables, respectively. Continuous variables and binary variables were compared using the Wilcoxon test. A P value <0.05 was considered to represent statistical significance.
Results
Patients’ characteristics
A total of 36 patients were enrolled and included in this retrospective study. Pathology revealed all cases were adenocarcinoma (100%, 36/36). The most common EGFR activating alterations were 19del (50%, 18/36) and L858R (47.22%, 17/36). Most patients (97.22%, 35/36) received first/second-generation EGFR-TKIs as first-line therapy. We performed mutation profiling analysis on 36 plasma samples of NSCLC lung adenocarcinoma and targeted 556 cancer-related genes using next-generation sequencing. EGFR T790M mutation was present in the plasma samples of 13 (36.11%) patients, obtained prior to third-generation EGFR-TKI treatment. In addition, 19 (52.78%) patients were EGFR 19 deletion-positive, 20 (55.56%) were EGFR 21 L858R mutation-positive, and 17 (47.22%) patients had multiple EGFR mutations detected prior to further treatment. The clinical characteristics of the 36 patients included in the retrospective analysis are shown in Table 1.
Gene alterations
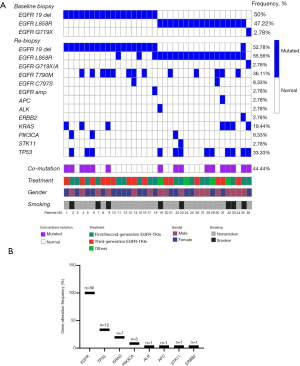
The EGFR sensitive mutations 19del or 21L858R were detected in 35 patients, and only 1 patient had the G719X/A mutation. The types of EGFR mutation detected by NGS were consistent with the results of the first biopsy, and only four patients had both the 19del and 21L858R mutations (Figure 1A). Multiple EGFR mutations were detected in 17 patients (47.22%), 13 of whom had complex exon T790M mutation and 4 of whom had more than one EGFR sensitive exon 19 or 21 mutation. EGFR amplification (2.78%, 1/36) and C797S mutation (5.56%, 2/36) were also found. Concomitant mutations were present in 44.4% (16/36) of the patients, and the most frequent accompanying somatic mutation was TP53 (33.3%, 12/36), followed by KRAS (19.44%, 7/36) and PIK3CA (8.33%, 3/36) (Figure 1B). Also, the presence of concomitant mutations was found to be significantly related to a history of smoking [87.5% (7 of 8) vs. 32.14% (9 of 28); Pearson chi-square, P=0.005].
Treatment outcome
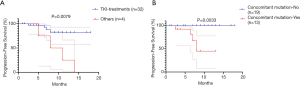
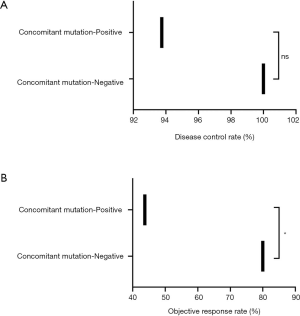
Of the 32 patients who continued to receive EGFR-TKI treatment, 15 received third-generation TKI, including 13 patients with positive EGFR T790M mutations. The patients who received EGFR-TKI treatment had significantly longer PFS than those who received other treatments [Figure 2A; P=0.0079, HR, 5.24 (95% CI, 0.79–34.69)]. Moreover,Of the 32 patients receiving EGFR-TKI treatment, those with concomitant genetic alterations showed a worse response (Figure 2B; P=0.0033). Patients with T790M mutations achieved longer PFS than EGFR T790M mutation-negative patients [Table 2, P=0.075; HR, 0.197 (95% CI: 0.05–0.75)]. Furthermore, patients who received third-generation EGFR TKI treatment presented a better outcome than those who received other treatments [Figure 3; HR, 5.98 (95% CI, 1.60–22.41); P=0.0468]. In addition, patients with concomitant mutations were significantly associated with poorer PFS [Table 2, P<0.001; HR, 18.27 (95% CI, 4.42–75.58)]. However, no statistically significant difference was found when sex, a history of smoking, treatment type, EGFR T790M mutation, and concomitant mutations were entered into the multivariable Cox proportional hazards regression model) (Table 2). Moreover, the presence of concomitant mutations was significantly associated with a lower objective response rate (43.75% vs. 80.0%; P=0.024), but not disease control rate (Figure 4).

Full table

Discussion
Molecular screening is crucial in choosing suitable EGFR-TKI therapy for patients with EGFR activating mutations. As a number of studies have shown, the rapid development of gene sequencing technology has opened the door for the broad application of dynamic monitoring of tumor driver genes in tumor diagnosis and treatment, including in identifying tumor genome maps, monitoring treatment responses, detecting early drug resistance, and predicting prognosis (19). This study retrospectively analyzed data from 36 patients with lung adenocarcinoma who were diagnosed with EGFR mutation after receiving first-line EGFR-TKI. The dynamic monitoring of tumor genomic profiles was performed on plasma samples from the patients, and 556 cancer-related genes were targeted using next-generation sequencing before the patients underwent further TKI treatment. The results show that advanced NSCLC patients with EGFR mutation have extensive co-mutations supported previous report (20). Furthermore, genetic covariation was found to have a negative impact on the response and survival of patients who received first-line EGFR-TKI treatment, which may be attributed to bypass activation of survival signaling pathways or tumor heterogeneity.
Several previous studies have investigated the relationship between co-occurring genetic changes and response and survival (11,20,21). These studies challenge the single driver oncogene perspective and reveal the potential function of concomitant genetic changes as an auxiliary driver of tumor progression. In the present study, 44.4% of patients harbored concomitant mutations, which were significantly associated with poorer objective response rate and PFS. The result is consistent with previous studies, suggesting that concomitant driver gene mutations play an important role in tumor progression and drug resistance, and may be a very important factor affecting clinical outcomes. However, in multivariate analysis, concomitant mutations were not significantly associated with PFS, which indicates that the adverse treatment results caused by concomitant mutations may be interfered by other factors. Interestingly, we also found a significantly higher incidence of concomitant mutations in patients who had a history of smoking. Moreover, the presence of the EGFR T790M mutation after first-generation EGFR TKI treatment was also significantly associated with greater efficacy of third-generation EGFR TKIs (the data was not shown), which provides a rationale for the superiority of third-generation EGFR TKI therapy over other types of therapy and the significance of molecular diagnosis in NSCLC progressive disease. Similar to the T790M mutation, C797S appears in EGFR exon 20, which determines that cysteine is replaced by serine at position 797. Several authors have documented that the appearance of C797S with negative T790M could maintain sensitivity to EGFR inhibitors, but not in T790M-positive cells in a preclinical setting (22,23). In our study, two patients had C797S accompanied by the T790M mutation, which suggests a possible correlation with poor response to third-generation TKI therapy. In addition, the activation of known by-pass signaling pathways as a mechanism of resistance was represented in our cohort by one patient who had STK11 mutation, three patients who had KRAS mutation, and one patient who had ERBB2 amplification. STK11 mutation presented in our study in only one patient, which has been reported as a mediator of the cold tumor immune microenvironment and a major driver in primary resistance to PD-1 axis inhibitors in non-squamous lung adenocarcinoma (24). KRAS activating mutations appear in patients treated with first-generation EGFR-TKIs and are considered to be a potential mechanism for EGFR-TKIs (25). Inhibition of EGFR by osimertinib may functionally deplete the carcinogenic EGFR signal to a level that can cause KRAS mutant cells to appear (26,27). Concurrent ERBB2 (HER2) is a rare, independent predictor of shorter progression time with EGFR TKI treatment. Owing to their clinical availability, HER2 inhibitors may be particularly clinically feasible. In the specimens obtained after EGFR TKI resistance, the copy number changes of ERBB2 were also enriched (28).
Our primary and subsequent biopsies, using tumor tissue and blood-derived ctDNA, respectively, detected, and identified potential actionable changes, particularly in relation to EGFR-sensitive mutations. Exon 18 G719X mutation is a less common EGFR-TKIs sensitizing mutation. It occurred in only one patient in our study (1/36, 2.8%), which was similar to the reported 3% incidence of EGFR mutations (29). Aside from four patients with 19del and 21 L858R co-mutations, there appeared to be high consistency between tissue ARMS PCR and ctDNA NGS. Compared with traditional detection methods, NGS technology is more efficient and has higher throughput for through tissue and blood samples, which can simultaneously detect point mutations, insertions, rearrangements and copy number mutations, and mutation abundance, and perform quantitative detection. It also saves financial resources and time in large-scale sequencing. Conventionally, tissue biopsies have been used to assess the mechanisms of acquired drug resistance (30). However, since re-biopsy is an invasive procedure, it is not always feasible and accepted by patients, and it cannot explain the heterogeneity of drug resistance mechanisms in different tumor sites of the same patient, so it has some limitations. Moreover, blood-derived ctDNA, also known as liquid biopsy, was a non-invasive option. Molecular analysis of the plasma collected during the onset of disease can provide timely monitoring of the disease response. Emerging studies have enhanced the prognostic significance of plasmatic mutational analysis in third-generation EGFR-TKIs candidate patients (31-33). Thus it is suggested that liquid biopsy technique is a powerful complement to tissue specimens and deserves further in-depth analysis, especially in first-line setting perspective.
This study has several limitations. First, the sample size was too small. Second, patients who received different lines of TKI treatment were included in the analysis. Third, data from our next-generation sequencing hotspots (excluding some other alterations, copy number changes, or chromosomal abnormalities) may have caused concomitant mutations to be underestimated.
In conclusion, the findings of this study emphasize the importance of using multiple molecular profiles. Concomitant genetic alterations were significantly associated with response to EGFR targeted therapy in patients with NSCLC, and may serve as an important negative predictor of TKI treatment response in patients with EGFR sensitive mutations. Therefore, research on multi-drug or sequential therapy must be urgently carried out to address the covariation that drives drug resistance.
Acknowledgments
Funding: The study was supported by the Competitive Distribution Projects of Zhanjiang Science and Technology Development Special Fund (2018A010220), and sponsored by Shanghai Tongshu Biotechnology Co., Ltd.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-679
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-679
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-679). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of the Affiliated Hospital of Guangdong Medical University (No. YJ2017062-1). Informed consent was obtained from all patients who underwent blood genetic analysis. All procedures performed in this study were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Liu Y, Xiong ZC, Sun X, et al. Impact of apatinib in combination with osimertinib on EGFR T790M-positive lung adenocarcinoma. Transl Cancer Res 2019;8:2151-63. [Crossref]
- Pao W, Miller V, Zakowski M, et al. EGF receptor gene mutations are common in lung cancers from "never smokers" and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A 2004;101:13306-11. [Crossref] [PubMed]
- Leonetti A, Facchinetti F, Tiseo M. Upfront osimertinib in EGFR-mutated non-small cell lung cancer: is brain still a sanctuary? Ann Transl Med 2018;6:S110. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Chen J, Soudy H. Osimertinib in the treatment of leptomeningeal disease in T790M-negative, epidermal growth factor receptor-mutated non-small cell lung cancer: a case report. Chin Clin Oncol 2019;8:29. [Crossref] [PubMed]
- Zhou Q, Zhang XC, Chen ZH, et al. Relative abundance of EGFR mutations predicts benefit from gefitinib treatment for advanced non-small-cell lung cancer. J Clin Oncol 2011;29:3316-21. [Crossref] [PubMed]
- Yang X, Zhuo M, Ye X, et al. Quantification of mutant alleles in circulating tumor DNA can predict survival in lung cancer. Oncotarget 2016;7:20810-24. [Crossref] [PubMed]
- Riely GJ, Pao W, Pham D, et al. Clinical course of patients with non-small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res 2006;12:839-44. [Crossref] [PubMed]
- Jackman DM, Yeap BY, Sequist LV, et al. Exon 19 deletion mutations of epidermal growth factor receptor are associated with prolonged survival in non-small cell lung cancer patients treated with gefitinib or erlotinib. Clin Cancer Res 2006;12:3908-14. [Crossref] [PubMed]
- Blakely CM, Watkins TBK, Wu W, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet 2017;49:1693-704. [Crossref] [PubMed]
- VanderLaan PA, Rangachari D, Mockus SM, et al. Mutations in TP53, PIK3CA, PTEN and other genes in EGFR mutated lung cancers: Correlation with clinical outcomes. Lung Cancer 2017;106:17-21. [Crossref] [PubMed]
- Canale M, Petracci E, Delmonte A, et al. Impact of TP53 Mutations on Outcome in EGFR-Mutated Patients Treated with First-Line Tyrosine Kinase Inhibitors. Clin Cancer Res 2017;23:2195-202. [Crossref] [PubMed]
- Jiang T, Zhao J, Zhao C, et al. Dynamic Monitoring and Predictive Value of Circulating Tumor Cells in EGFR-Mutated Advanced Non-Small-Cell Lung Cancer Patients Treated With First-Line EGFR Tyrosine Kinase Inhibitors. Clin Lung Cancer 2019;20:124-33.e2. [Crossref] [PubMed]
- Imamura F, Uchida J, Kukita Y, et al. Monitoring of treatment responses and clonal evolution of tumor cells by circulating tumor DNA of heterogeneous mutant EGFR genes in lung cancer. Lung Cancer 2016;94:68-73. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491-8. [Crossref] [PubMed]
- Lai Z, Markovets A, Ahdesmaki M, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res 2016;44:e108. [Crossref] [PubMed]
- Hofman P. The challenges of evaluating predictive biomarkers using small biopsy tissue samples and liquid biopsies from non-small cell lung cancer patients. J Thorac Dis 2019;11:S57-64. [Crossref] [PubMed]
- Hong S, Gao F, Fu S, et al. Concomitant Genetic Alterations With Response to Treatment and Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Patients With EGFR-Mutant Advanced Non-Small Cell Lung Cancer. JAMA Oncol 2018;4:739-42. [Crossref] [PubMed]
- Wang Z, Cheng Y, An T, et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): a phase 2, single-arm, multicentre clinical trial. Lancet Respir Med 2018;6:681-90. [Crossref] [PubMed]
- Ercan D, Choi HG, Yun CH, et al. EGFR Mutations and Resistance to Irreversible Pyrimidine-Based EGFR Inhibitors. Clin Cancer Res 2015;21:3913-23. [Crossref] [PubMed]
- Niederst MJ, Hu H, Mulvey HE, et al. The Allelic Context of the C797S Mutation Acquired upon Treatment with Third-Generation EGFR Inhibitors Impacts Sensitivity to Subsequent Treatment Strategies. Clin Cancer Res 2015;21:3924-33. [Crossref] [PubMed]
- Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov 2018;8:822-35. [Crossref] [PubMed]
- Del Re M, Tiseo M, Bordi P, et al. Contribution of KRAS mutations and c.2369C > T (p.T790M) EGFR to acquired resistance to EGFR-TKIs in EGFR mutant NSCLC: a study on circulating tumor DNA. Oncotarget 2017;8:13611-9. [Crossref] [PubMed]
- Hata AN, Niederst MJ, Archibald HL, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med 2016;22:262-9. [Crossref] [PubMed]
- Unni AM, Lockwood WW, Zejnullahu K, et al. Evidence that synthetic lethality underlies the mutual exclusivity of oncogenic KRAS and EGFR mutations in lung adenocarcinoma. Elife 2015;4:e06907. [Crossref] [PubMed]
- Yu HA, Suzawa K, Jordan E, et al. Concurrent Alterations in EGFR-Mutant Lung Cancers Associated with Resistance to EGFR Kinase Inhibitors and Characterization of MTOR as a Mediator of Resistance. Clin Cancer Res 2018;24:3108-18. [Crossref] [PubMed]
- Kobayashi S, Canepa HM, Bailey AS, et al. Compound EGFR mutations and response to EGFR tyrosine kinase inhibitors. J Thorac Oncol 2013;8:45-51. [Crossref] [PubMed]
- Planchard D, Popat S, Kerr K, et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2018;29:iv192-iv237. [Crossref]
- Lin CC, Shih JY, Yu CJ, et al. Outcomes in patients with non-small-cell lung cancer and acquired Thr790Met mutation treated with osimertinib: a genomic study. Lancet Respir Med 2018;6:107-16. [Crossref] [PubMed]
- Wu YL, Sequist LV, Hu CP, et al. EGFR mutation detection in circulating cell-free DNA of lung adenocarcinoma patients: analysis of LUX-Lung 3 and 6. Br J Cancer 2017;116:175-85. [Crossref] [PubMed]
- Bordi P, Del Re M, Minari R, et al. From the beginning to resistance: Study of plasma monitoring and resistance mechanisms in a cohort of patients treated with osimertinib for advanced T790M-positive NSCLC. Lung Cancer 2019;131:78-85. [Crossref] [PubMed]


