PI3K-AKT-mTOR pathway alterations in advanced NSCLC patients after progression on EGFR-TKI and clinical response to EGFR-TKI plus everolimus combination therapy
Introduction
Lung cancer is the most frequently diagnosed cancers and the leading cause of cancer-related deaths (1). The identification of activating oncogenic aberrations such as EGFR mutations has significantly revolutionized the therapeutic routines for non-small cell lung cancer (NSCLC) patients. Epidermal growth factor receptor tyrosine kinase inhibitors (EGFR TKIs) are associated with superior efficacy in NSCLC patients with activating EGFR mutations (2-5). Despite dramatic clinical response achieved, patients treated with EGFR TKIs still inevitably develop acquired resistance. Several resistant mechanisms have been reported, including secondary mutations in EGFR, histologic transformation, as well as bypass or alternative pathway activation (6,7).
PI3K-AKT-mTOR pathway is frequently altered in cancers, involved in the regulation of multiple cellular functions including cell proliferation, survival, differentiation, adhesion, motility and invasion. PI3K-AKT-mTOR pathway activation plays an important role in EGFR-TKI resistant events (8-10). Clinical studies have demonstrated that EGFR mutant patients with PI3K pathway activation achieved shorter progression free survival (PFS) and worse overall survival (OS) (11-13). Alterations of PI3K-AKT-mTOR pathway can occur through activation of tyrosine kinase receptors upstream of PI3K, PIK3CA amplification as well as mutations in KRAS, PI3K, AKT, TSC1 and TSC2, or loss of PTEN (14,15). To date, it remains a challenge to overcome PI3K-pathway associated resistance and improve clinical benefit from EGFR TKIs.
Nowadays, only two mTOR inhibitors, everolimus and temsirolimus, have been approved for cancer treatment. Everolimus is a mTOR inhibitor targeting mTORC1 and has been approved for clinical use in numerous cancers including renal cell carcinoma, breast cancer and pancreatic neuroendocrine tumors (16-19). Preclinical studies have demonstrated that everolimus could overcome resistance to EGFR inhibitors and produce a cooperative effect with EGFR inhibitors in several human cancer cell lines resistant to EGFR inhibitors (20). In vitro studies also suggested the clinical efficacy of combining mTOR inhibitors with EGFR-TKIs in TKIs-resistant NSCLC cell lines (21,22). Tolerable toxicity and disease control of combination therapy have been reported in NSCLC patients carrying EGFR mutations (23,24). Although subsequent clinical trials showed limited effect of combination therapy in unselected NSCLC patients, the potential effect in patients with specific genetic aberrations still entails further exploration (25,26).
There are limited studies about the prevalence of activated PI3K pathway in EGFR-TKI resistant patients. Herein, we analyzed comprehensive molecular profiling in 605 patients with EGFR-TKI treatment history and explored the co-occurrence of common mutations through PI3K-AKT-mTOR pathway in patients with acquired resistance to EGFR TKIs. Furthermore, six patients with a history of everolimus plus EGFR TKIs treatment were retrospectively collected to estimate the antitumor activity and safety of the tragedy.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-141).
Methods
Patients
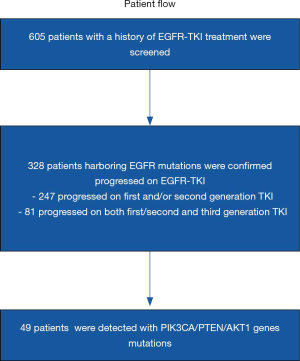
A total of 605 NSCLC patients with EGFR-TKI treatment history who were referred to OrigiMed (Shanghai, China) for next generation sequencing (NGS) testing between September 2016 and October 2018 were involved. Genetic information and clinical characteristics such as gender, histology, treatment process was documented at the time of referral. Based on these data, 328 patients were further collected to analyze the co-occurrence of common mutations along PI3K-AKT-mTOR pathway after progression on EGFR TKIs (Figure 1). For these patients, Formalin fixed and paraffin embedded (FFPE) tumor or blood samples were collected at the onset of EGFR TKI treatment progression for NGS to estimate potential resistance mechanisms. Furthermore, six patients with specific mutations were retrospectively included to evaluate the antitumor activity and safety of EGFR TKI plus everolimus therapy. Patient data were collected through digital medical records. Written consents were provided by all patients.
This study was conducted in accordance with the Declaration of Helsinki Declaration (as revised in 2013) and approved by Sun Yat-sen University Cancer Center IRB (B2018-092-01).
Genetic analysis
NGS was performed by FFPE tissues or circulating tumor DNA (ctDNA) in the laboratory of OrigiMed. The number of genes tested on tumor tissue panel ranged from 37 to 450. Diluted libraries were sequenced with a mean coverage of 900× for FFPE samples and 300× for matched blood samples on an Illumina NextSeq-500 Platform (Illumina Incorporated, San Diego, CA). For ctDNA assays, the number of genes tested ranged from 18 to 329. Diluted libraries were sequenced to a mean coverage of 3,000×. Genomic data was then analyzed based on 18 cancer-related genes covered by all panels. Common molecular mechanisms of EGFR TKI resistance, including T790M mutation, MET amplification, ERBB2 amplification and BRAF mutations were documented. PIK3CA, AKT1 mutations and PTEN loss, the most universal mutations leading to activation of PI3K-AKT-mTOR pathway and covered by all the NGS examinations were recorded as well.
Response evaluation
Six EGFR mutant NSCLC patients with specific mutations along PI3K-AKT-mTOR pathway, were treated with EGFR TKIs and everolimus. Computed tomography (CT) on chest and abdomen were routinely performed at the first month after initiation of combination therapy and then once eight weeks later. Cerebral magnetic resonance imaging (MRI) were carried out once the patients was diagnosed with brain metastasis. Response assessments were based on Response Evaluation Criteria in Solid Tumors (RECIST) (27). PFS was defined as the interval from the date of treatment commencement to the date of treatment failure or death.
Statistical analysis
All the statistical analyses included in this study were analyzed by R software. A P value <0.05 was considered statistically significant.
Results
Clinical and general genetic data from patients
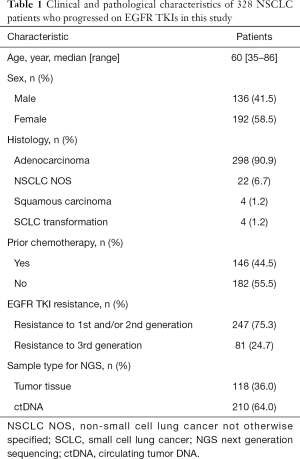
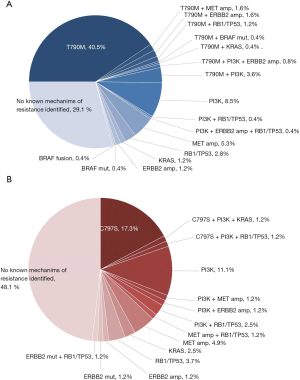
Among 605 advanced NSCLC patients reviewed, 328 patients were confirmed progressed on EGFR-TKIs, including 298 adenocarcinoma, 22 NSCLC not otherwise specified (NOS), 4 squamous carcinoma and 4 SCLC transformation. The median age was 60 (range, 35–86) years and 58.5% (192/328) were female (Table 1). According to clinical information generated, 81 patients were progressed on third generation EGFR-TKI, and 247 patients on earlier generation TKIs. The resistance mechanisms of EGFR-TKI treatment revealed by NGS were shown in Figure 2. EGFR T790M, PI3K activation, MET amplification, KRAS, ERBB2 amplification, BRAF mutations, BRAF fusion alone and complex mutations were identified 40.5%, 8.5%, 5.3%, 1.2%, 1.2%, 0.4%, 0.4% and 10.4%, respectively among patients with acquired resistance to first and/or second generation TKIs. Unknown resistance mechanisms were observed in 29.1% of these cases (Figure 2A). As for patients resistant to osimertinib, 17.3%, 11.1%, 4.9%, 2.5%, 1.2%, 9.7%, 48.1% were conformed emergence of C797S, PI3K activation, MET amplification, KRAS, ERBB2 amplification, complicate mutations and unclear mechanisms (Figure 2B). Clinical characteristics between patients with PI3K pathway alterations and without PI3K pathway alterations were compared and no significant difference were observed in age, sex, histology (Table S1).
Full table

Full table
PI3K-AKT-mTOR pathway alterations
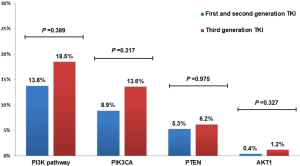
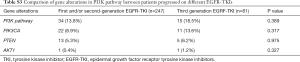
Forty-nine (14.9%) patients with EGFR-TKI resistant were identified emergence of PIK3CA, PTEN and AKT1, involving 45 adenocarcinomas and 4 NSCLC-NOS patients. Such mutations were detected in 13.8% (34/247) and 18.5% (15/81) patients who had developed resistance to earlier generation EGFR TKIs or third generation EGFR TKI, respectively (Figure 3).

PIK3CA, PTEN and AKT1 variations were detected in 33 (10.1%), 18 (5.5%) and 2 (0.6%) of EGFR TKI resistant patients respectively. Three patients had both PIK3CA and PTEN mutations. Hotspot of PIK3CA mutations included E545, H1047 and E542, accounting for 60.5% of PIK3CA mutations (Figure 4). No significant differences were found in PI3K-AKT-mTOR pathway alterations among groups with different EGFR mutations (EGFR exon19 deletion mutations/EGFR L858R/uncommon EGFR mutations, see Table S2). There was also no significant difference in PI3K-AKT-mTOR pathway alterations between patients resistant to earlier generation EGFR TKIs and osimertinib (Figure S1, Table S3). Of note, PI3K pathway genetic mutations occurred with T790M in 11 patients resistant to first and/or second-generation EGFR TKIs, which might influence the efficacy of osimertinib. Except for EGFR mutations, the most common mutations co-existed with PI3K pathway were TP53 (57.1%) and RB1 (10.2%) (Figure 3).


Full table
Full table
Response to EGFR-TKI plus everolimus treatment
Six patients with EGFR-TKI and everolimus history were retrospectively collected to assess the effect and safety of EGFR TKI plus everolimus treatment. Clinical data including gender, age and treatment history as well as genetic information were shown in Table 2. All patients were stage IV NSCLC including 5 females with adenocarcinoma and 1 male with squamous carcinoma. The median age is 68 years old. Except patient 3, none of them has a smoking history. All patients have received at least one line of standard therapy before combination treatment. Four patients started combination therapy as fourth-line therapy (Table 2). Five patients used to receive osimertinib monotherapy prior to combination therapy, three of whom were EGFR T790M positive prior to osimertinib therapy. EGFR T790M were undetected in these patients after progression on osimertinib (Table S4). Acquired PIK3CA mutations, PTEN loss were detected after progression on EGFR TKIs in all patients, with patient 4 harboring both PIK3CA mutation and PTEN loss. Four patients were treated with osimertinib 80 mg and the rest gefitinib 250 mg. The dose of everolimus was 5 mg daily in all patients. According to RECISIT 1.1, six patients achieved stable disease (SD). One patient didn’t acquire disease control. The median PFS of 2.1 months (95% confidence interval, 1.35–4.3 months, range, 0.9–4.4 months). Treatment-related adverse events encompassed oral ulcer, rash (Table 2). One patent switched to other treatment due to intolerable grade III oral ulcer and one patient passed away under worsening condition. Most patients experienced progressive disease (PD) eventually.

Full table

Full table
Patient 1 received everolimus plus osimertinib and achieved SD with a PFS of 4.2 months. The patient transferred to chemotherapy due to lung disease progression. For patient 2, although pulmonary stability was observed through CT after one-month of combination treatment, enlarged lesions in liver occurred after 3 months, resulting in PD. On receiving osimertinib plus everolimus, patient 3 experienced shrinkage in pulmonary mass and pleural lesions. Unfortunately, the patient passed away after 4.4 months because of respiratory failure. Patient 4 achieved SD after one month, while discontinued treatment for intolerable oral ulcer. Although patient 5 acquired SD, we cannot obtain detained information after she left the hospital last time and died. PFS was calculated until the last date she came to hospital. No disease control was observed in patient 6.
Discussion
PI3K-AKT-mTOR pathway activation has been implicated as an important resistance mechanism in patients treated with EGFR TKIs. Co-occurrence of PI3K-related mutations with EGFR activating mutations predicts worse prognosis and shorter PFS with EGFR TKI treatment. Through retrospective analysis on NGS data collected from OrigiMed, we clarified the popularity of the most common PI3K-related mutations in EGFR-TKI resistant patients. Furthermore, through retrospective study of six NSCLC patients, we found that EGFR TKIs pus everolimus, one of the few clinical available mTOR inhibitors (everolimus and temsirolimus), might not be enough to overcome EGFR TKI resistance induced by abnormal PI3K pathway activation.
According to OrigiMed NGS data, we found that PI3K pathway alterations represent 14.9% in EGFR TKI resistance events, serving as a common resistant mechanism. PIK3CA, PTEN and AKT1 variations were detected in 33 (10.1%), 18 (5.5%) and 2 (0.6%) of patients respectively. PI3K activation caused by three aberrations were detected in 13.4% (34/247) and 18.5% (15/81) patients who had developed resistance to earlier generation EGFR TKIs or third generation EGFR TKI, respectively. No significant differences were observed in PI3K-AKT-mTOR pathway alterations among patients with different EGFR mutations and among patients progressed on different EGFR TKIs. Notably, PI3K pathway gene mutations occurred in 11 patients with EGFR T790M, which may influence the efficacy of osimertinib. To the best of our knowledge, our study initially revealed the distribution of common mutations involving PI3K activation in patients resistant to EGFR TKIs, which were difficult to overcome.
With advancements of precision medicine and fast-emerging molecular sequencing technology, increasing researches focus on co-targeting therapies (28-30). Despite that five patients (83.3%) in our cohort achieved disease control, the median PFS was short and most patients experienced PD ultimately. There are several potential reasons implicated in the failure of combination therapy. First, everolimus was an allosteric mTOR inhibitor specifically targets the mTORC1 complex, not mTORC2. Inhibition of mTORC1 alone can activate the mTORC1 negative feed-back loop, resulting in activation of AKT via S6K dependent upregulation of the IRS-1 and TGFR-1 pathways (31). Therefore, the inhibition of mTORC1 is unable to suppress the PI3K pathway completely. Agents like dual Pan-PI3K and mTOR inhibitors, or mTOR kinase inhibitors, which encompass a wide range of molecular targets of PI3K-AKT-mTOR pathway might offer more favorable activity (32). Second, PI3K pathway activation interacts with other signaling pathway like MAPK pathway at many levels, resulting in failure (33,34). Enhanced clinical response might be achieved by application of PI3K inhibition sensitizer such as CDK4/6 inhibition, or co-targeting other resistant pathways which interacts with PI3K pathway (35-37). Third, intolerable toxicity prevents the use of the combination therapy, with grade 3 AEs reported in two patients and treatment discontinuation in one patient. Currently, safety and toxicity profile of PI3K pathway inhibitors remains problematic for further clinical applying (38). Novel PI3K inhibitors like isoform-selective-PI3K inhibitors against specific biomarkers have fewer off-target toxicities and require development (39). Adverse effects might be alleviated through optimization of dosing schedules such as intermittent dosing rather than concomitant dosing (40). To sum up, novel selective-compounds targeting specific aberration proteins, improving therapeutic efficacy and safety profile with appropriate treatment schedules are required. Corresponding genetic biomarkers predictive of selective-agents effect entails urgent exploration. Last, multiple mutations co-existed with PI3K-associated aberrations. Recent studies have found that increasing burden of co-occurrence mutations was associated with worse prognosis against EGFR TKIs (12,41). The existence of TP53 and RB1 mutations also have been reported as negative factors for clinical benefit (12,42,43). In this retrospective cohort, most samples tested by large-panel NGS carried several concurrent mutations besides EGFR and PI3K pathway aberrations, and carries concomitant TP53 mutations or RB1 mutations, which might lead to the complexity of resistance mechanisms and worse prognosis. Hence, co-targeting EGFR mutations and PI3K pathway was insufficient in such patients. Precision combination therapies targeting multiple aberrations or multi-targeted agents demands development.
Several limitations also exist in our study. The sample size of patients receiving EGFR TKIs plus everolimus is small and might not represent actual efficacy in all patients. Moreover, combination therapies were treated as late-line treatment in most retrospective cases, causing complicated to observe real efficacy. We only analyzed the most common mutations, PI3KCA, PTEN and AKT1 mutations, because these three aberrations were covered by the NGS with the least panels. Other less common mutations involving PI3K-AKT-mTOR pathway weren’t taken into consideration. Therefore, a larger cohort with wide-range NGS are needed to determine a more accurate distribution of PI3K-AKT-mTOR mutations in EGFR mutant patients.
In conclusion, our study revealed that PIK3CA pathway was activated in at least 14.9% of EGFR-TKI resistant patients. Combination therapy of EGFR-TKIs and everolimus revealed limited antitumor activity in NSCLC patients with dysregulation of PI3K-AKT-mTOR pathway. Further studies on better strategies are imperative to optimize clinical outcomes.
Acknowledgments
We thank the patients and their families in the study.
Funding: This work was supported by National Key R&D Program of China (2016YFC0905500, 2016YFC0905503), Chinese National Natural Science Foundation Project (81972556, 81772476, 81602005, 81872499, and 81702283), Science and Technology Program of Guangdong (2017B020227001), Science and Technology Program of Guangzhou (201607020031, 201704020072), CSCO-Hengrui Cancer Research Fund (Y-HR2016-113).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-141
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-141
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-141). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki Declaration (as revised in 2013) and approved by Sun Yat-sen University Cancer Center IRB (B2018-092-01). Written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013;31:3327-34. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Lee DH. Treatments for EGFR-mutant non-small cell lung cancer (NSCLC): The road to a success, paved with failures. Pharmacol Ther 2017;174:1-21. [Crossref] [PubMed]
- Tan CS, Gilligan D, Pacey S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol 2015;16:e447-e459. [Crossref] [PubMed]
- Fumarola C, Bonelli MA, Petronini PG, et al. Targeting PI3K/AKT/mTOR pathway in non small cell lung cancer. Biochem Pharmacol 2014;90:197-207. [Crossref] [PubMed]
- Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol 2010;28:1075-83. [Crossref] [PubMed]
- Zhang Y, Kwok-Shing Ng P, Kucherlapati M, et al. A Pan-Cancer Proteogenomic Atlas of PI3K/AKT/mTOR Pathway Alterations. Cancer Cell 2017;31:820-832.e3. [Crossref] [PubMed]
- Kim HR, Cho BC, Shim HS, et al. Prediction for response duration to epidermal growth factor receptor-tyrosine kinase inhibitors in EGFR mutated never smoker lung adenocarcinoma. Lung Cancer 2014;83:374-82. [Crossref] [PubMed]
- Kim Y, Lee B, Shim JH, et al. Concurrent Genetic Alterations Predict the Progression to Target Therapy in EGFR-Mutated Advanced NSCLC. J Thorac Oncol 2019;14:193-202. [Crossref] [PubMed]
- Blakely CM, Watkins TBK, Wu W, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet 2017;49:1693-704. [Crossref] [PubMed]
- Lee YR, Chen M, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat Rev Mol Cell Biol 2018;19:547-62. [Crossref] [PubMed]
- Lui VW, Hedberg ML, Li H, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov 2013;3:761-9. [Crossref] [PubMed]
- FDA Approves Drug Combo for Kidney Cancer. Cancer Discov 2016;6:687-8.
- Papadimitrakopoulou V. Development of PI3K/AKT/mTOR pathway inhibitors and their application in personalized therapy for non-small-cell lung cancer. J Thorac Oncol 2012;7:1315-26. [Crossref] [PubMed]
- Baselga J, Campone M, Piccart M, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med 2012;366:520-9. [Crossref] [PubMed]
- Yao JC, Shah MH, Ito T, et al. Everolimus for advanced pancreatic neuroendocrine tumors. N Engl J Med 2011;364:514-23. [Crossref] [PubMed]
- Bianco R, Garofalo S, Rosa R, et al. Inhibition of mTOR pathway by everolimus cooperates with EGFR inhibitors in human tumours sensitive and resistant to anti-EGFR drugs. Br J Cancer 2008;98:923-30. [Crossref] [PubMed]
- La Monica S, Galetti M, Alfieri RR, et al. Everolimus restores gefitinib sensitivity in resistant non-small cell lung cancer cell lines. Biochem Pharmacol 2009;78:460-8. [Crossref] [PubMed]
- Dong S, Zhang XC, Cheng H, et al. Everolimus synergizes with gefitinib in non-small-cell lung cancer cell lines resistant to epidermal growth factor receptor tyrosine kinase inhibitors. Cancer Chemother Pharmacol 2012;70:707-16. [Crossref] [PubMed]
- Milton DT, Riely GJ, Azzoli CG, et al. Phase 1 trial of everolimus and gefitinib in patients with advanced nonsmall-cell lung cancer. Cancer 2007;110:599-605. [Crossref] [PubMed]
- Papadimitrakopoulou VA, Soria JC, Jappe A, et al. Everolimus and erlotinib as second- or third-line therapy in patients with advanced non-small-cell lung cancer. J Thorac Oncol 2012;7:1594-601. [Crossref] [PubMed]
- Price KA, Azzoli CG, Krug LM, et al. Phase II trial of gefitinib and everolimus in advanced non-small cell lung cancer. J Thorac Oncol 2010;5:1623-9. [Crossref] [PubMed]
- Besse B, Leighl N, Bennouna J, et al. Phase II study of everolimus-erlotinib in previously treated patients with advanced non-small-cell lung cancer. Ann Oncol 2014;25:409-15. [Crossref] [PubMed]
- Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer 2017;17:637-58. [Crossref] [PubMed]
- Vilar E, Perez-Garcia J, Tabernero J. Pushing the envelope in the mTOR pathway: the second generation of inhibitors. Mol Cancer Ther 2011;10:395-403. [Crossref] [PubMed]
- Janku F, Yap TA, Meric-Bernstam F. Targeting the PI3K pathway in cancer: are we making headway? Nat Rev Clin Oncol 2018;15:273-91. [Crossref] [PubMed]
- Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov 2014;13:140-56. [Crossref] [PubMed]
- Dogruluk T, Tsang YH, Espitia M, et al. Identification of Variant-Specific Functions of PIK3CA by Rapid Phenotyping of Rare Mutations. Cancer Res 2015;75:5341-54. [Crossref] [PubMed]
- Bosch A, Li Z, Bergamaschi A, et al. PI3K inhibition results in enhanced estrogen receptor function and dependence in hormone receptor-positive breast cancer. Sci Transl Med 2015;7:283ra51. [Crossref] [PubMed]
- Vora SR, Juric D, Kim N, et al. CDK 4/6 inhibitors sensitize PIK3CA mutant breast cancer to PI3K inhibitors. Cancer Cell 2014;26:136-49. [Crossref] [PubMed]
- Muranen T, Meric-Bernstam F, Mills GB. Promising rationally derived combination therapy with PI3K and CDK4/6 inhibitors. Cancer Cell 2014;26:7-9. [Crossref] [PubMed]
- Esposito A, Viale G, Curigliano G. Safety, Tolerability, and Management of Toxic Effects of Phosphatidylinositol 3-Kinase Inhibitor Treatment in Patients With Cancer: A Review. JAMA Oncol 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Rodon J, Dienstmann R, Serra V, et al. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat Rev Clin Oncol 2013;10:143-53. [Crossref] [PubMed]
- Hoeflich KP, Merchant M, Orr C, et al. Intermittent administration of MEK inhibitor GDC-0973 plus PI3K inhibitor GDC-0941 triggers robust apoptosis and tumor growth inhibition. Cancer Res 2012;72:210-9. [Crossref] [PubMed]
- Hong S, Gao F, Fu S, et al. Concomitant Genetic Alterations With Response to Treatment and Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Patients With EGFR-Mutant Advanced Non-Small Cell Lung Cancer. JAMA Oncol 2018;4:739-42. [Crossref] [PubMed]
- Canale M, Petracci E, Delmonte A, et al. Impact of TP53 Mutations on Outcome in EGFR-Mutated Patients Treated with First-Line Tyrosine Kinase Inhibitors. Clin Cancer Res 2017;23:2195-202. [Crossref] [PubMed]
- VanderLaan PA, Rangachari D, Mockus SM, et al. Mutations in TP53, PIK3CA, PTEN and other genes in EGFR mutated lung cancers: Correlation with clinical outcomes. Lung Cancer 2017;106:17-21. [Crossref] [PubMed]