
Genomic characteristics in Chinese non-small cell lung cancer patients and its value in prediction of postoperative prognosis
Introduction
There are about 733,300 new diagnosed lung cancer in China per year and also ranks as the leading cause of cancer-related mortality in China, with non-small cell lung cancer (NSCLC) as the predominant subtype (1). Comprehensive molecular profiling of NSCLC was revolutionized by the development of next-generation sequencing (NGS) and highlights the importance of molecular classification for NSCLC. Previous genomic studies using NGS revealed different gene mutation landscapes between Caucasian and Asian patients with lung cancer (2-5). However, most of genomics profiling studies of lung cancer with large sample size have predominantly conducted in Caucasians. Large studies in Chinese NSCLC patients are still needed (4,6-8).
The TNM classification by the American Joint Committee on Cancer represents the standard prognostic system for early lung cancer (9). However, despite recent improvements in the staging, prognosis varies considerably within the same TNM stage. It has been reported that several key clinical factors such as gender, age, histology, and molecular indicators are better predictors than TNM stage for prognosis (10-12). Nomogram, based on univariate analysis and multivariate Cox stepwise regression model, was established to predict and quantify survival probability by generating crucial parameters which contribute to prognosis risk. In several cancer types, nomograms based on multivariate models were more precise predictors of survival than classical TNM staging system alone (13,14). A Chinese study generated a nomogram using clinical parameters for survival prediction in NSCLC after surgery and this model displayed significantly superior performance than TNM staging alone (15). Since some genetic abnormalities also have prognostic value, integration of clinical and molecular characteristics has the potential of improving the ability to predict prognosis.
Here we aimed to elucidate the comprehensive genomic characteristics of Chinese resected NSCLC patients using a panel of 295 cancer-related genes by NGS. Moreover, we established nomogram model to provide integrated predictors consisting of clinical and genomic characteristics to predict postoperative prognosis and improve risk stratification. We present the following article in accordance with the STROBE Reporting Checklist (available at http://dx.doi.org/10.21037/tlcr-19-664).
Methods
Patients
Tumor samples were collected from 511 treatment-naive lung cancer patients (without any other primary tumors) who underwent surgical resection at Tianjin Medical University Cancer Institute & Hospital between May 2009 and November 2012. The number of lung cancer patients undergoing surgery during that period determined the sample size. This study was approved by a central ethic committee of Tianjin Medical University Cancer Institute & Hospital (No. E2016060A). The clinical trial registration number was NCT03609918. Staging was according to the 8th edition tumor, node, metastasis (TNM) criteria (9). Histological classification was assessed according to the latest World Health Organization criteria (16). OS was calculated from the surgery date to death or last follow-up. 29 patients were lost to follow-up who were excluded from survival analysis.
NGS library preparation and sequencing
Capture-based targeted ultra-deep sequencing was performed on 511 resected tumors (including 484 frozen tissue and 27 FFPE samples) by the OncoScreen panel, which spans 2.02 MB of human genome and consists of all exons and critical introns of 295 genes (The list of genes was provides in Table S1). DNA was extracted using the QIAamp DNA FFPE tissue Kit (Qiagen) according to the manufacturer's instructions. DNA was sheared, end repaired, phosphorylated before adaptor ligation. The ligated fragments with size of 200–400 bp were selected by magnetic beads hybridized with probe baits and amplified by PCR. Indexed samples were sequenced on a NextSeq 500 (Illumina, Inc., USA) with pair-end reads.
Full table
The sequencing data in the FASTQ format were mapped to the human genome (hg19) using BWA aligner 0.7.10. Local alignment optimization, variant calling and annotation were performed using GATK 3.2, MuTect, and VarScan, respectively. Variants were filtered using the VarScan filter pipeline, with loci with depth <100X filtered out. At least 5 supporting reads were needed for indels in tissue samples, while 8 supporting reads were needed for SNVs to be called. CNVs were analyzed with in-house algorithm based on sequencing depth of coverage data of capture intervals. The minimum threshold of copy number gain or loss was CN >2.75 or CN <1.75 for hotspot genes, and CN >3 or CN <1.5 for others. DNA translocations were analyzed using FACTERA.
Construction of nomogram
Each clinical and molecular factor was evaluated in a univariate Cox proportional hazard model for OS outcomes. Variables with P-value of less than 0.1 were selected for the following multivariate analysis using Stepwise algorithm in variable selection procedure. A nomogram was established on the basis of multivariate analysis results and conducted using R (version 3.3.1) with package rms. For each patient, each variable was assigned to a point on a scale of 0 to 100, and the calculated total score by summing the points corresponded to the probability of 3-year or 5-year OS.
Kaplan–Meier survival curves were constructed after dividing patients into two groups based on median risk score. The performance of the nomogram was evaluated through its calibration and discrimination. Calibration indicates the concordance of predicted survival and actual survival, whereas discrimination refers to the ability of a nomogram model to stratify patients with different outcomes and is represented by Harrell’s concordance index (C-index) (17). The C-index value is from 0.5 to 1.0. A C-index of 0.5 indicates concordance by chance, and a C-index of 1.0 represents a prefect discriminative ability. Utilizing 10X cross validation to validate the nomogram model.
Statistical analysis
Fisher’s exact test was conducted to compare the mutational frequency of lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) between our cohort and TCGA population, as well as to analyze the concurrence and mutual exclusivity between genes. Risk group stratification based on nomogram prediction was delineated using Kaplan-Meier survival curve and log-rank test was used to compare the difference between survival groups. The C-indexes of different nomogram models were compared using ANOVA. A P-value less than 0.05 was considered statistically significant.
Results
Clinical and demographic characteristics
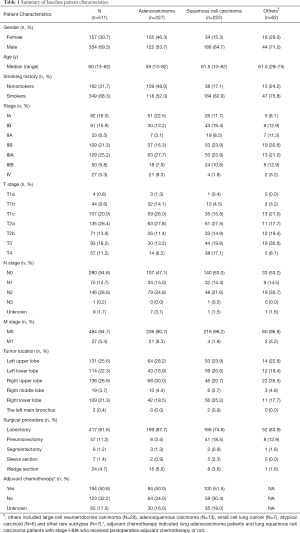
In this cohort, 30.7% (157/511) were females and 69.3% (354/511) were males. Patients had a median age of 60 years at surgery, ranging from 13 to 82 years. There were 68.3% of patients (349/511) with smoking history and 31.7% (162/511) non-smokers. Among all, 44.4% (227/511) patients were diagnosed as LUAD while 43.4% (222/511) were classified as LUSC. Other histology subtype constituted the rest 12.1% (62/511) of the cohort. Patients with stage I, II and IIIA accounted for 31.9% (163/511), 27.8% (142/511) and 25.2% (129/511), respectively. Clinico-pathologic characteristics of this cohort are summarized in Table 1.
Full table
Overview of baseline mutation spectrum
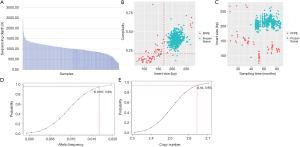
The mean coverage depth was 1,040× (Figure S1A) among all samples. Frozen tissues (N=484) displayed higher mean insert size and library complexity than FFPE samples (N=27), which indicates less DNA degradation and higher DNA quality (Figure S1B). The length of insert size was inversely correlated with sample collection time in FFPE samples (P<0.0001) but not in frozen tissues (Figure S1C). Limits of detection for single-nucleotide variations (SNV) and insertion/deletion (indel), translocation/fusion, copy number amplification (CNA) were 1.67% (Figure S1D), 2% and 3 (Figure S1E), respectively.
Among the 511 samples, 98.6% had at least one genetic aberrance detected. A total of 5,245 somatic mutations spanning 294 genes were identified, consisting of 4,059 SNVs, 549 indels, 607 CNAs, and 30 translocations. 7 patients had no mutation identified. Overall mutation spectrum of this cohort is demonstrated in Figure 1A.
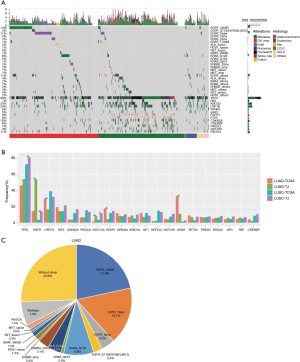
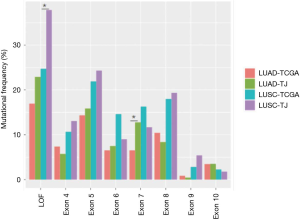
We compared mutation frequency of LUAD and LUSC between this Chinese cohort and TCGA population (http://www.cbioportal.org) (Figure 1B). The most frequently mutated gene was TP53, and Chinese LUSC cohort, but not LUAD, displayed a higher rate of TP53 alterations than that of TCGA population (LUAD, 53.7% vs. 46.1%, P=0.220; LUSC, 81.1% vs. 72.3%, P=0.042; Fisher’s exact test). We also observed that Chinese LUSC patients harbored more TP53 loss of function (LOF) mutations than TCGA (P=0.007), and Chinese LUAD patients had more TP53 exon 7 mutations than TCGA cohort (P=0.038, Figure S2). As a well-known druggable target, EGFR aberrances ranked as the predominant mutations among all oncogenic drivers and were identified in 53.7% of Chinese LUAD patients, compared to 14.4% in TCGA (P<0.001). KRAS mutations occurred in 11.0% of Chinese LUAD patients, whereas displayed a prevalence of 32.6% in the TCGA database (P<0.001). In addition, we found that the mutation frequencies of KEAP1 (4.4% vs. 17.4%, P<0.001) and LRP1B (16.3% vs. 29.6%, P<0.001) in Chinese LUAD cohort were significantly lower than in TCGA. The mutation rate of NOTCH3 (LUAD: 8.4% vs. 1.3%, P<0.001; LUSC: 11.3% vs. 3.4%, P=0.004) was higher for both LUAD and LUSC in this Chinese cohort than in TCGA.
Overall, 74.4% (169/227) of Chinese LUAD patients had at least one druggable mutation detected, including EGFR (L858R, 19del, S768I, G719X, L861Q, 20ins), ERBB2 (20ins, amplification), MET (exon 14 skipping and amplification), BRAF (V600E), PIK3CA, ROS1, RET and ALK rearrangement (Figure 1C).
Mutually exclusive and co-occurring patterns of mutations
Pairwise mutual exclusivity and co-occurrence analysis were performed separately on LUAD (N=227) and LUSC (N=222) and the analysis revealed distinct patterns between these two NSCLC subtypes (Figure 2A,B).
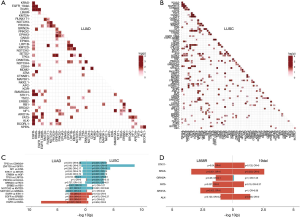
In LUAD, EGFR displayed significant exclusivity with ALK (OR =0.10, P=0.001), KRAS (OR =0, P<0.001), STK11 (OR =0.10, P=0.001) and ERBB2 (OR =0.10, P=0.001) (Figure 2C), consistent with previous reports (18,19). Pairs with significant co-occurrence identified in LUAD included NOTCH3/GRIN2A (OR =9.30, P<0.001), NOTCH2/KMT2D (OR =9.10, P=0.001), ERBB2/RB1 (OR>10, P=0.001), and TP53/LRP1B (OR =2.70, P=0.012). In LUSC patients, eight co-occurring gene pairs were identified, including GRIN2A/FAT3 (OR =4.61, P=0.001), PIK3CA/KLHL6 (OR =5.13, P=0.001), TP53/LRP1B (OR =5.60, P<0.001), ERBB4/HGF (OR >10, P=0.001), STK11/MTOR (OR>10, P<0.001), NF1/ATM (OR>10, P<0.001), ZNF703/ FGFR1 (OR>10, P<0.001), and TP53/CDKN2A (OR>10, P<0.001). No mutually exclusive gene pair was identified. Interestingly, TP53/LRP1B displayed co-occurrence relationship in both LUAD and LUSC. Taken together, these observations indicated that the mutual exclusivity/co-occurrence pattern is mainly unique to specific histological subtype, suggesting unique tumorigenesis mechanisms in the different subtypes.
Since it has been reported that patients harboring EGFR exon 19 deletions (19del) or L858R display different drug sensitivity and clinical outcomes in NSCLC (20,21), we interrogated the distinct underlying exclusivity and co-occurrence of the two variants in LUAD. EGFR L858R and 19del both displayed strong mutual exclusivity with KRAS (OR =0, P<0.001; OR =0, P=0.003), whereas ARID1A (OR =0, P=0.003), FAT3 (OR =0, P=0.027) and STK11 (OR =0, P=0.040) showed mutual exclusivity only to L858R and ALK (OR =0, P=0.045) was significantly mutually exclusive with 19del (Figure 2D).
Clinical relevance of mutated pathways
We investigated the distribution of mutations across Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in different histological subtype clinical subgroups, including gender, age, smoking status, tumor location, tumor size and tumor stage (Figure 3). Among LUAD patients, mutations in VEGF and NOTCH signaling pathways commonly occurred in male (P=0.043 and 0.003), while HIF1 signaling pathway were more prone to mutate in female (P=0.047). Age was positively correlated with gene alterations involved in apoptosis, P53 pathway, cell cycle, PI3K/AKT signaling pathway and central carbon metabolism (P=0.017, 0.006, 0.024, 0.009 and 0.014) in LUSC, but not in LUAD. Our analysis also revealed that patients with smoking history accumulated more mutations in VEGF signaling pathway, apoptosis and NOTCH pathway in LUAD (P=0.003, 0.001 and 0.045), but more gene alterations in mTOR signaling pathway in LUSC (P=0.044).
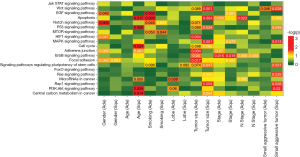
Next, we interrogated the correlation of tumor features (location and TNM stage) and involved pathways. As to tumor location, upper lobe lesions were associated with mutations in cancer microRNAs in LUAD (P=0.038). In terms of TNM stage, we found that mutations in HIF1, adherents junction, ERBB2 and focal adhesion pathway were enriched in LUAD with small tumor size (P=0.004, 0.004, 0.040 and 0.017); while mutations in WNT, apoptosis, P53, MAPK, pluripotency of stem cells, and RAP1 pathways in small-sized LUSC (P=0.001, 0.004, 0.034, 0.001, 0.047 and 0.009). Mutations in ERBB2 pathway showed negative correlation with stage both in LUAD and LUSC (P=0.016 and 0.019). And mutations in apoptosis pathway negatively correlated with N stage in LUAD patients (P=0.023). We also observed that small and aggressive tumors (T1–2, N >0, M >0) accumulated more mutations in WNT pathway, P53, MAPK, pluripotency of stem cells, RAS, RAP1, and PI3K/AKT pathways (P=0.028, 0.048, 0.013, 0.031, 0.029, 0.028 and 0.02) in LUSC, while had few mutations in WNT pathway in LUAD (P=0.046). Some clinical parameters displayed different associations with pathways in LUAD and LUSC, suggesting a histology-specific mechanism for driving pathogenesis in lung cancers.
Prognostic nomogram for survival prediction in NSCLC
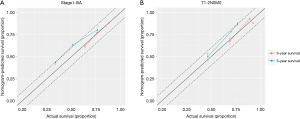
Nomograms combining both clinical and genomic characteristics were developed to predict postoperative prognosis in surgically resected NSCLC. Seven independent prognostic factors were selected and entered into the nomogram based on cox multivariable analyses. The nomogram for 3- and 5-year OS prediction (N=360) demonstrated that wt-EPHA3 and late stage showed the largest contribution to inferior OS, followed by mut-KRAS, wt-ETV5, mut-ALK and old age (Figure 4A). Histology type only slightly contributed to the model. The calibration plot displayed a good correlation between predicted survival and actual outcomes (Figure S3A). The discriminative ability of the nomogram model was also assessed using the Harrell’s C-index. The C-index of established nomograms for survival prediction consisting of both clinical and genomic characteristics was 0.663 (95% CI, 0.638–0.688), which was higher than that of clinical features only (0.624, 95% CI, 0.599–0.649; 0.663 vs. 0.624, P=0.0001) or considering stage only (0.618, 95% CI, 0.593–0.643; 0.663 vs. 0.618, P<0.0001). The C-index was internally validated by performing 10×cross validation. The corrected C-index for OS was 0.646.
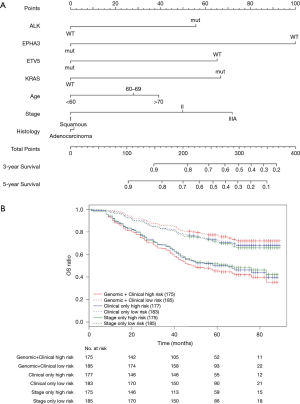
Next, we performed log-rank analysis to further demonstrate the nomogram performance in stratifying prognostic risks based on the nomogram “score”. The median follow-up time was 51.5 months. The high- and low-risk subgroups divided by total score from nomogram model including both clinical and genomic characteristics had a median OS of 49 months (95% CI, 42–76 months) and not reached (NR) (95% CI, NR–NR). The median OS for the two subgroups sorted by total score from nomograms with clinical features were 65 months (95% CI, 45–NR) and NR (95% CI, NR–NR). The high- and low-risk subgroups divided by stage had median OS of 65 months (95% CI, 45–NR months) and NR (95% CI, NR–NR). Patients with different prognostic risk were more significantly separated by nomogram model including both clinical and genomic characteristics (HR =2.939, 95% CI, 2.070–4.173) than by nomograms with clinical features (HR =2.171, 95% CI, 1.547–3.045) or stage only (HR =1.976, 95% CI, 1.414–2.761) (2.939 vs. 2.171, P<0.001; 2.939 vs. 1.976, P<0.001, Figure 4B). Overall, in this cohort, nomogram of multivariate setting consisting of combined clinical and genomic characteristics exhibited reasonable discrimination and provided more precise prediction for individualized clinical outcomes than predictions based on clinical features or staging only.
Prognostic nomogram in T1-2N0M0 sub-cohort
It is recognized that prognostic markers could provide basis for developing personalized approaches to improve the survival of early-stage NSCLC patients (10-12). Therefore, we constructed nomograms specifically within the stage T1-2N0M0 sub-group to identify significant prognostic parameters to predict OS in these clinically low-risk patients. Nomogram model established for 3- and 5-year OS prediction revealed that mut-KRAS, mut-TP53_exon 8, older age and LUSC histology were correlated with inferior OS (N=165, Figure 5A). The calibration plots presented good agreement between nomogram prediction and actual survival for 3-year/5-year OS (Figure S3B). The discrimination of nomograms presented by C-index was 0.681 (95% CI, 0.639–0.723) for OS prediction, which was significantly higher than that of clinical features only (0.629, 95% CI, 0.587–0.671; 0.681 vs. 0.629, P=0.0026) or stage only (0.594, 95% CI, 0.558–0.635; 0.681 vs. 0.594, P<0.0001). The corrected C-index for OS was 0.655 with 10-fold internal cross validation.
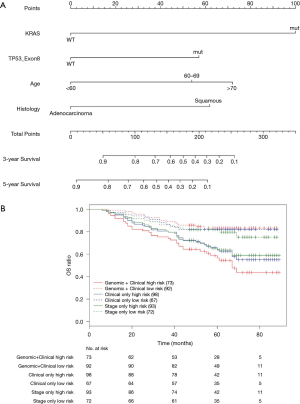
After stratifying patients into different risk sub-groups based on predicted total scores, each sub-group significantly presented a distinct prognosis to each other. The stratification based on nomogram with integrated clinical and genomic characteristics (median OS, high vs. low: 67 (95% CI, 57–NR) vs. NR (95% CI, NR–NR); HR =3.499) showed more significant difference than that of clinical features (median OS, high vs. low: NR (95% CI, 67–NR) vs. NR (95% CI, NR–NR); HR =2.327) or stage only (median OS, high vs. low: NR (95% CI, 67-NR) vs. NR (95% CI, NR–NR); HR =1.861) (3.499 vs. 2.327, P=0.002; 3.499 vs. 1.861, P<0.001, Figure 5B), indicating a potential of more precise risk stratification than traditional TNM staging system in early-stage patients.
Discussion
In the present study, we demonstrated differential mutational distributions in multiple key genes in Chinese NSCLC compared to the TCGA population, and revealed unique mutual exclusivity and co-occurrence features in LUAD and LUSC. Moreover, we established nomogram models with integrated clinical and genomic characteristics to provide more precise postoperative prognostic prediction than traditional TNM staging system in resectable NSCLC, and specifically in the clinically low-risk population.
As the most frequently mutated gene, TP53 had higher mutation rate in LUSC in this cohort than TCGA. Asian lung adenocarcinoma patients harbor more frequent EGFR mutations than Caucasians (22), whereas KRAS is more prone to occur in Caucasians than Asians. The results in our study were congruent with previous studies (4). However, our study showed that the frequencies of less common mutations such as LRP1B, KEAP1 and NOTCH3 were different from TCGA data. Ethnicity could be associated with the status of gene mutations because very few Asians were included in the TCGA database. Additionally, smoking status was significantly different in the two populations (non-smokers percentage in TCGA cohort and our cohort were 13.4% and 31.7%, respectively), which may also contribute to the different mutation frequencies of these genes. Further extensive genomic investigations will help to better elucidate underlying genetic difference between ethnic sub-groups.
We explored the mutual exclusivity/co-occurrence of genomic alterations from different driver pathways in specific NSCLC subtype, which will be helpful to understand molecular mechanisms of oncogenesis. It was reported that mutual exclusivity is commonly seen within one specific pathway but not across pathways (23,24). Whereas, genes were more prone to display co-occurrence relationship across several pathway pairs (25,26). This pattern was also observed in Chinese cohort. For example, EGFR were exclusive with ALK, KRAS and ERBB2, all of which were involved in MAPK pathways activation, indicating that mutations in exclusive gene pairs may be independently sufficient for regulation of downstream oncogenic signaling. Moreover, we observed alterations in pathways were remarkably related to clinical features. LUAD and LUSC showed different mutated pathways in the same clinical subgroups. The observations suggest that LUAD and LUSC have diverse underlying mechanisms of tumorigenesis or tumor maintenance.
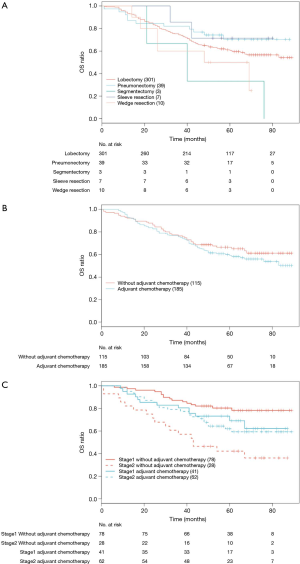
Nomograms have been proven to be a convenient and reliable algorithmic approach to predict prognosis by evaluating potential important factors (27-29). Limited studies have developed nomograms for predicting survival in patients with resected lung cancer (15,30-32). To the best of our knowledge, most studies focused on clinical characteristics associated variables for survival estimation. In this study, we integrated factors of both gene signature and clinical characteristics to establish models for prognosis prediction. Stage and age were consistently identified as important markers for survival prediction both in previous studies and our work. While surgical procedure and adjuvant chemotherapy might be important to prognosis prediction in resectable NSCLC, survival analysis showed that surgical procedure had no effect on OS in our study (P=0.104, Figure S4A). A univariate Cox proportional hazard model for OS outcomes indicated that adjuvant chemotherapy had no significant effect on OS (P=0.290, Figure S4B). Actually, after stratifying by stage, survival analysis showed that only stage II patients benefited from adjuvant chemotherapy, and stage I patients did not benefit from adjuvant chemotherapy in our cohort (stage II P=0.033, stage I P=0.158, Figure S4C). The multivariate Cox analysis which included histology, age and stage showed that adjuvant chemotherapy was not associated with OS (HR =0.810, P=0.379). The reason of no correlation between adjuvant chemotherapy and OS could be attributed to the high percentage of stage I patients in this cohort (N=119, 39.7%). Whether they can derive benefits from adjuvant therapy is still debatable (33-35). Therefore, adjuvant chemotherapy was not included in the prognostic nomogram models.
As regard to gene signature for survival estimation, there is still a controversy over the prognostic value of KRAS mutation in early-stage resectable NSCLC patients. Several studies demonstrated that KRAS mutation was associated with poorer clinical outcome (36-38), whereas some investigators reported only modest or even no prognostic effects of KRAS mutation (39-41). In our study, KRAS mutation was found to be an indicator of inferior prognosis for OS in early-stage NSCLC. Furthermore, we derived several other gene signatures which can be used to predict survival in our cohorts. Our results suggested the importance of risk stratification based on combination of clinical characteristics and gene alterations in early-stage NSCLC for more precise survival prediction.
There were several limitations in our analysis. Firstly, the post-surgery treatment could be a confounding factor. Secondly, the model of postoperative prognosis we developed has not been validated through external dataset, so its application for other patients should be performed cautiously. Additional prognostic signature explorations are still needed to further validate and improve our model.
Conclusions
In summary, this comprehensive genomic profiling revealed Chinese NSCLC patients have unique gene profile, and distinct mutual exclusivity and co-occurrence patterns existed between LUAD and LUSC. The combination of genomic with clinical characteristics showed more accurate prediction of postoperative prognosis in early-stage NSCLC, indicating the importance of developing the next TNM staging system that integrates genomic data with clinical characteristics.
Acknowledgments
We presented an interim analysis of this study as oral presentation at the IASLC 19th World Conference on Lung Cancer in Toronto, Canada, September 22–26, 2018.
Funding: This work was supported by National Key Research and Development Program of China Grant (grant number 2016YFC0905501, 2016YFC0905500); National Natural Science Foundation of China (grant number 81772484, 81772488, 81672304); Tianjin Thousand Talents program; Tianjin Cancer Hospital Clinical Trial Project (grant number C1705); and AstraZeneca Investment (China) Co., Ltd.
Footnote
Reporting Checklist: The authors have completed the STROBE Reporting Checklist. Available at http://dx.doi.org/10.21037/tlcr-19-664
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-19-664
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-19-664). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by a Central Ethic Committee of Tianjin Medical University Cancer Institute & Hospital (No. E2016060A). Informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Li S, Choi YL, Gong Z, et al. Comprehensive Characterization of Oncogenic Drivers in Asian Lung Adenocarcinoma. J Thorac Oncol 2016;11:2129-40. [Crossref] [PubMed]
- Kim Y, Hammerman PS, Kim J, et al. Integrative and comparative genomic analysis of lung squamous cell carcinomas in East Asian patients. J Clin Oncol 2014;32:121-8. [Crossref] [PubMed]
- Liu L, Liu J, Shao D, et al. Comprehensive genomic profiling of lung cancer using a validated panel to explore therapeutic targets in East Asian patients. Cancer Sci 2017;108:2487-94. [Crossref] [PubMed]
- Wen S, Dai L, Wang L, et al. Genomic Signature of Driver Genes Identified by Target Next-Generation Sequencing in Chinese Non-Small Cell Lung Cancer. Oncologist 2019;24:e1070-81. [Crossref] [PubMed]
- Shen H, Zhu M, Wang C. Precision oncology of lung cancer: genetic and genomic differences in Chinese population. NPJ Precis Oncol 2019;3:14. [Crossref] [PubMed]
- Goldstraw P, Chansky K, Crowley J, et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J Thorac Oncol 2016;11:39-51. [Crossref] [PubMed]
- Chansky K, Sculier JP, Crowley JJ, et al. The International Association for the Study of Lung Cancer Staging Project: prognostic factors and pathologic TNM stage in surgically managed non-small cell lung cancer. J Thorac Oncol 2009;4:792-801. [Crossref] [PubMed]
- Kawaguchi T, Takada M, Kubo A, et al. Performance status and smoking status are independent favorable prognostic factors for survival in non-small cell lung cancer: a comprehensive analysis of 26,957 patients with NSCLC. J Thorac Oncol 2010;5:620-30. [Crossref] [PubMed]
- Sculier JP, Chansky K, Crowley JJ, et al. The impact of additional prognostic factors on survival and their relationship with the anatomical extent of disease expressed by the 6th Edition of the TNM Classification of Malignant Tumors and the proposals for the 7th Edition. J Thorac Oncol 2008;3:457-66.
- Zaak D, Burger M, Otto W, et al. Predicting individual outcomes after radical cystectomy: an external validation of current nomograms. BJU Int 2010;106:342-8. [Crossref] [PubMed]
- Wang Y, Li J, Xia Y, et al. Prognostic nomogram for intrahepatic cholangiocarcinoma after partial hepatectomy. J Clin Oncol 2013;31:1188-95. [Crossref] [PubMed]
- Liang W, Zhang L, Jiang G, et al. Development and validation of a nomogram for predicting survival in patients with resected non-small-cell lung cancer. J Clin Oncol 2015;33:861-9. [Crossref] [PubMed]
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Iasonos A, Schrag D, Raj GV, et al. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol 2008;26:1364-70. [Crossref] [PubMed]
- Gainor JF, Varghese AM, Ou SH, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res 2013;19:4273-81. [Crossref] [PubMed]
- Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069-75. [Crossref] [PubMed]
- Sugio K, Uramoto H, Onitsuka T, et al. Prospective phase II study of gefitinib in non-small cell lung cancer with epidermal growth factor receptor gene mutations. Lung Cancer 2009;64:314-8. [Crossref] [PubMed]
- Tudor R, Kopciuk K, D'Silva A, et al. Addressing disease progression in EGFRmut+ NSCLC patients. Ann Oncol 2016;27:1252P-P.
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Yeang CH, McCormick F, Levine A. Combinatorial patterns of somatic gene mutations in cancer. FASEB J 2008;22:2605-22. [Crossref] [PubMed]
- Vandin F, Upfal E, Raphael BJ. De novo discovery of mutated driver pathways in cancer. Genome Res 2012;22:375-85. [Crossref] [PubMed]
- Leiserson MD, Vandin F, Wu HT, et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet 2015;47:106-14. [Crossref] [PubMed]
- Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nature Reviews Cancer 2019;19:495-509. [Crossref] [PubMed]
- Valentini V, van Stiphout RG, Lammering G, et al. Nomograms for predicting local recurrence, distant metastases, and overall survival for patients with locally advanced rectal cancer on the basis of European randomized clinical trials. J Clin Oncol 2011;29:3163-72. [Crossref] [PubMed]
- Han DS, Suh YS, Kong SH, et al. Nomogram predicting long-term survival after d2 gastrectomy for gastric cancer. J Clin Oncol 2012;30:3834-40. [Crossref] [PubMed]
- Karakiewicz PI, Briganti A, Chun FK, et al. Multi-institutional validation of a new renal cancer-specific survival nomogram. J Clin Oncol 2007;25:1316-22. [Crossref] [PubMed]
- Filosso PL, Guerrera F, Evangelista A, et al. Prognostic model of survival for typical bronchial carcinoid tumours: analysis of 1109 patients on behalf of the European Association of Thoracic Surgeons (ESTS) Neuroendocrine Tumours Working Group. Eur J Cardiothorac Surg 2015;48:441-7; discussion 447. [Crossref] [PubMed]
- Tanvetyanon T, Finley DJ, Fabian T, et al. Prognostic nomogram to predict survival after surgery for synchronous multiple lung cancers in multiple lobes. J Thorac Oncol 2015;10:338-45. [Crossref] [PubMed]
- Kent MS, Mandrekar SJ, Landreneau R, et al. A Nomogram to Predict Recurrence and Survival of High-Risk Patients Undergoing Sublobar Resection for Lung Cancer: An Analysis of a Multicenter Prospective Study (ACOSOG Z4032). Ann Thorac Surg 2016;102:239-46. [Crossref] [PubMed]
- Artal Cortés Á, Calera Urquizu L, et al. Adjuvant chemotherapy in non-small cell lung cancer: state-of-the-art. Transl Lung Cancer Res 2015;4:191-7. [PubMed]
- Morgensztern D, Samson PS, Waqar SN, et al. Early Mortality in Patients Undergoing Adjuvant Chemotherapy for Non-Small Cell Lung Cancer. J Thorac Oncol 2018;13:543-9. [Crossref] [PubMed]
- Wang J, Wu N, Lv C, et al. Should patients with stage IB non-small cell lung cancer receive adjuvant chemotherapy? A comparison of survival between the 8th and 7th editions of the AJCC TNM staging system for stage IB patients. J Cancer Res Clin Oncol 2019;145:463-9.
- Nelson HH, Christiani DC, Mark EJ, et al. Implications and prognostic value of K-ras mutation for early-stage lung cancer in women. J Natl Cancer Inst 1999;91:2032-8. [Crossref] [PubMed]
- Schabath MB, Welsh EA, Fulp WJ, et al. Differential association of STK11 and TP53 with KRAS mutation-associated gene expression, proliferation and immune surveillance in lung adenocarcinoma. Oncogene 2016;35:3209-16. [Crossref] [PubMed]
- Meng D, Yuan M, Li X, et al. Prognostic value of K-RAS mutations in patients with non-small cell lung cancer: a systematic review with meta-analysis. Lung Cancer 2013;81:1-10. [Crossref] [PubMed]
- Shepherd FA, Domerg C, Hainaut P, et al. Pooled analysis of the prognostic and predictive effects of KRAS mutation status and KRAS mutation subtype in early-stage resected non-small-cell lung cancer in four trials of adjuvant chemotherapy. J Clin Oncol 2013;31:2173-81. [Crossref] [PubMed]
- La Fleur L, Falk-Sorqvist E, Smeds P, et al. Mutation patterns in a population-based non-small cell lung cancer cohort and prognostic impact of concomitant mutations in KRAS and TP53 or STK11. Lung Cancer 2019;130:50-8. [Crossref] [PubMed]
- Zer A, Ding K, Lee SM, et al. Pooled Analysis of the Prognostic and Predictive Value of KRAS Mutation Status and Mutation Subtype in Patients with Non-Small Cell Lung Cancer Treated with Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors. J Thorac Oncol 2016;11:312-23. [Crossref] [PubMed]


