A multicenter real-world study of tumor-derived DNA from pleural effusion supernatant in genomic profiling of advanced lung cancer
Introduction
With the tremendous progress in the field of targeted therapy and immune-oncology, the landscape of advanced non-small lung cancer (NSCLC) therapies has rapidly been evolving beyond chemotherapy over the last few years. Targeted therapies against driver mutations such as epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), and ROS proto-oncogene 1 (ROS1) in patients with non-small cell lung cancer (NSCLC) have outperformed traditional chemotherapy in improving patients’ survival (1,2). Incorporating molecular genetic testing into standard clinical care is the first step to identify those patients. However, tumor tissues for molecular profiling are not always available from advanced patients. Therefore, the use of tumor-derived cell-free DNA (cfDNA) isolated from body fluids, including plasma, pleural effusion (PE), cerebrospinal fluids, saliva, and urine, for cancer genomic profiling is being investigated (3-6). Plasma derived cfDNA was widely accepted as a noninvasive option for tumor genomic profiling in patients with advanced stage malignancies, and also gained popularity for detecting tumor evolution and monitoring minimal residual disease in the era of precision medicine.
In advanced lung cancer patients, PE develops as a consequence of tumor cell invasion of the pleural cavity (7). As well as tumor cfDNA, the supernatant of PE contains floating malignant cells, caused by impaired pleural fluid drainage and abnormal plasma extravasation to the pleural space (8). PE supernatant is always collected for diagnostic and therapeutic purposes through thoracentesis. Previous studies have used the cellular sediments and/or supernatants of PE to detect EGFR mutations with Sanger sequencing, amplification refractory mutation system (ARMS) polymerase chain reaction (PCR), mutant-specific PCR, digital PCR, and next-generation sequencing (NGS) (9-15). Recently, genomic profiling using PE was investigated (16); however, the performance of genomic profiling using PE in a real-world setting has yet to be fully investigated. Moreover, as almost all previous studies focused on detecting actionable mutation, little is known for the use of PE in identifying resistant mutations of treated patients.
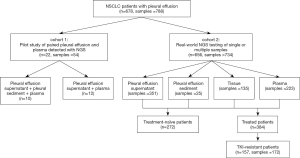
In this study, PE and plasma samples were simultaneously collected from 22 patients with advanced lung cancer (cohort 1) to evaluate the feasibility of using PE-derived DNA for clinical molecular diagnoses as a pilot study. And then in cohort 2, 732 samples from 656 NSCLC patients with pleural effusion who underwent NGS in a real-world setting were analyzed to verify the efficacy of pleural effusion in detecting actionable mutations and identifying resistance mechanisms of targeted therapy. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-882).
Methods
Patient recruitment
From September 2016 to December 2016, 22 lung cancer patients with pleural effusion were prospectively enrolled at the Department of Oncology, Sun Yat-sen University Cancer Center (Guangzhou, China) and the First Affiliated Hospital of Nanjing Medical University (Nanjing, China); these patients comprised study cohort 1. To analyze the real-world efficacy of pleural effusion in detecting actionable mutations, 656 NSCLC patients with pleural effusion who underwent NGS in a College of American Pathologists-accredited laboratory, Geneplus-Beijing (Beijing, China) from January 2017 to January 2020, were retrospectively analyzed as cohort 2. This study was conducted in accordance with the principles of the Declaration of Helsinki (as revised in 2013). The ethics committee of the First Affiliated Hospital of Nanjing Medical University (2019-SRFA-226) approved the study, and written informed consent was obtained from all patients.
DNA extraction, library preparation, and target enrichment
PE aliquots (10 mL) were centrifuged to separate the supernatant from the cell sediment. cfDNA from the PE supernatant and genomic DNA from cell sediment samples was extracted separately. All tissue samples included in this study underwent pathology review onsite to confirm histologic classification and the adequacy of the tumor tissues, which required a minimum of 20% of tumor cells. Genomic tumor DNA was extracted from the tumor tissues and PE sediments using the QIAamp DNA mini kit (Qiagen, Valencia, CA, USA). cfDNA was extracted from 4–5 mL of isolated plasma and PE supernatant using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Valencia, CA, USA). Genomic DNA was extracted from white blood cells as a germline control using the DNeasy Blood Kit (Qiagen, Valencia, CA, USA). DNA concentration was measured using a Qubit fluorometer and the Qubit dsDNA HS (High Sensitivity) Assay Kit (Invitrogen, Carlsbad, CA, USA). The size distribution of the cfDNA was assessed using an Agilent 2100 BioAnalyzer and a DNA HS kit (Agilent Technologies, Santa Clara, CA, USA). Sequencing libraries were prepared from ctDNA using KAPA DNA Library Preparation Kits (Kapa Biosystems, Wilmington, MA, USA), and genomic DNA sequencing libraries were prepared with Illumina TruSeq DNA Library Preparation Kits (Illumina, San Diego, CA, USA). In total, 786 libraries from 678 patients were hybridized to custom-designed biotinylated oligonucleotide probes (Roche NimbleGen, Madison, WI, USA), covering ~230 Kbp genomic regions of 59 genes in cohort 1 and ~1.4 Mbp genomic regions of 1,021 cancer-related genes in cohort 2 (http://fp.amegroups.cn/cms/bbdc4b478fe987717afb65e96c7dac3e/TLCR-20-882-1.pdf, http://fp.amegroups.cn/cms/875622dace17595c30f33d0815c8c2a3/TLCR-20-882-2.pdf) using the Illumina Nextseq CN 500 or Gene + Seq 2000 instrument (17,18).
Sequencing and data analysis
Sequencing data were analyzed using default parameters. Adaptor sequences and low-quality reads were removed. The clean reads were aligned to the reference human genome (hg19) using the Burrows-Wheeler Aligner (BWA; version 0.7.12-r1039). GATK (version 3.4-46-gbc02625) was employed for realignment and recalibration. Single nucleotide variants (SNV) were called using MuTect (version 1.1.4) and NChot, an in-house software developed for reviewing hotspot variants. Small insertions and deletions (InDel) were determined by GATK. CONTRA (v2.0.8) was used to identify somatic copy number alterations. All final candidate variants were manually verified with Integrative Genomics Viewer. Targeted capture sequencing required a minimal mean effective depth of coverage of 300× and 1,000× in tissues and plasma samples, respectively. Targetable genomic alterations simultaneously detected by this assay included base substitutions, short insertions and deletions, focal gene amplifications and homozygous deletions (copy number alterations), and select gene fusions and rearrangements.
Statistical analyses
All the statistical analyses were performed using GraphPad Prism (v. 8.0; GraphPad Software, La Jolla, CA, USA) software. Associations between any two categorical variables were analyzed with Fisher’s exact test. A two-sided P value of <0.05 represented statistical significance.
Results
Study design and patient demographics
A total of 678 patients (51.4%, male) with stage IV non-small cell lung cancer who had PE at diagnosis or during disease progression were enrolled (Table 1). The median age at diagnosis was 61 (range, 25–94) years. Of the patients, 254 (37.5%) had distant metastasis (M1b/M1c stage), and 424 (62.5%) did not (M1a stage). There were 629 (92.8%) cases of adenocarcinoma, 36 (5.3%) cases of squamous carcinoma, 5 (0.7%) cases of adenosquamous carcinoma, 3 cases of other rare subtypes, and 5 (1.2%) cases of NSCLC without histological classification after NGS testing.

Full table
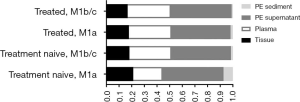
PE and matched plasma samples from 22 patients (cohort 1) were collected simultaneously as a pilot study. To verify the real-world efficacy of PE in genomic profiling, 734 samples from 656 consecutive patients with PE (cohort 2) were retrospectively analyzed. In the real-world setting, the majority of patients (86.7%, 569/656) had a single PE, plasma or tissue sample tested, while 87 patients had multiple samples tested simultaneously or consequently. In total, 351 PE supernatant samples, 224 plasma samples, 138 tissue samples, and 25 PE sediment samples were collected in cohort 2 (Figure 1). Among the 656 NSCLC patients in cohort 2, 413 were M1a stage, with tumors located only in the chest, while 243 were M1b/M1c stage, with 1 or more distant metastases. Moreover, 272 patients were newly diagnosed and 384 had previously been treated. So these 734 samples were divided into 4 groups: the treatment-naive M1a group (n=244); the treatment-naive M1b/c group (n=66), the treated M1a group (n=217); and the treated M1b/c group (n=207). Comparisons between the 4 groups revealed PE supernatant as the preferred choice for all the patients: 46.6–48.2% using PE supernatant, 23.3–34.8% using plasma, 16.8–21.2% using tissue and 0.96–7.3% of PE sediment). Moreover, tissue samples were used slightly more often in the treatment-naive M1a stage group than in the other groups (21.2% vs. 16.8–18.2%). Interestingly, the treatment-naive M1a stage group also had the largest proportion of PE sediment samples (Figure 2).
PE supernatant in pilot study
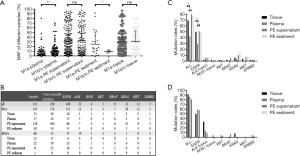
In cohort 1, the PE samples of 10 patients were separated into supernatant and cell sediment fractions for independent extraction of cfDNA. So, 10 samples of PE sediment, 22 samples of PE supernatant, and 22 plasma cfDNA samples were prepared for NGS. Among them, 90.1% (20/22) of PE supernatant samples, 90% (9/10) of PE sediment samples, and 95.4% (21/22) of plasma cfDNA samples had detectable somatic alterations. Actionable EGFR, ALK, ROS1, KRAS, ERBB2, MET, RET, and BRAF mutations were detected in 18 of the 22 supernatant samples and 10 of the plasma samples (18/22 vs. 10/22, P=0.01, Figure 3A). Three additional ALK fusions, 3 EGFR EX19 del mutations, 1 ROS1 fusion, and 1 KRAS G12D mutation were detected in the PE supernatant samples. For the 10 patients with PE sediment samples, both of the 2 ALK fusions were missed in the sediment samples, while plasma missed 1 of the 2 ALK fusions, 1 of the 3 KRAS mutations, and 1 of the 5 EGFR mutations detected by PE supernatant. Then, the maximal somatic allele frequency (MSAF) of PE-cfDNA was compared to those of supernatant from the 22 patients. The MSAF of the supernatant was significantly higher than that of the matched plasma cfDNA (P=0.01) (Figure 3B). We speculated that the lower MSAF in plasma may account for the inferior detection efficacy of PE sediment and plasma compared to PE supernatant.
PE supernatant in a real-world setting
To further analyze the efficacy of PE supernatant in the real-world setting, 656 NSCLC patients with pleural effusion in cohort 2 were retrospectively analyzed. Among them, 413 were M1a stage while 243 were M1b/M1c stage. Targeted NGS of 1,021 cancer-relevant genes was applied for genomic profiling of cohort 2. The table (http://fp.amegroups.cn/cms/ea9ba561f7823c93a42e9c8ef64a84bb/TLCR-20-882-3.pdf) provides a detailed list of somatic alterations identified in each sample from the patients. To avoid potential disturbance of treatment on MSAF, only MSAFs of different samples from the treatment naive patients were compared for further study. As expected, the MSAF of plasma in M1a stage patients was significantly lower than that in M1b/c stage patients (4.4%±9.6% vs. 9.0%±14.1%, P<0.01), but the MSAFs of PE supernatant and tissue were similar between the M1a and M1b/c stage patients (30.0%±29.4% vs. 32.8%±30.27%, P=0.52, Figure 4A). Therefore, we compared actionable EGFR, ALK, ROS1, KRAS, ERBB2, MET, RET, and BRAF mutations in treatment-naive patients (n=272) separately for the M1a and M1b/c groups (Figure 4B). The detection rate of actionable EGFR, ALK, ROS1, RET, BRAF, KRAS, MET, and ERBB2 mutations in supernatant was comparable to that in tissue samples reported previously (19). However, PE supernatant demonstrated higher sensitivity than plasma in detecting actionable mutations in M1a disease (84.7% for PE supernatant vs. 42.1% for plasma, P<0.01), especially for EGFR (50.8% for PE supernatant vs. 29.8% for plasma, P<0.01) (Figure 4C). However, in M1b/c stage patients, PE-supernatant and plasma had similar sensitivity in detecting actionable mutations (80.7% for PE-supernatant vs. 86.4% for plasma (P=0.58) (Figure 4D). This indicated that MSAF may account for the detection efficacy of different sample types.
PE supernatant for resistant mutations
Among the 384 NSCLC patients who had previously received treatment, 157 were confirmed by NGS as resistant to either first-, second, or third-generation EGFR-TKIs, or crizotinib, which is a TKI for ALK or ROS1 fusion. From these 157 patients, 94 PE supernatant samples, 54 plasma samples, 20 tissue samples, and 4 sediment samples were tested to explore the underlying resistance mechanisms. Of the samples, 117 were resistant to first- or second-generation EGFR-TKIs, 42 samples were resistant to osimertinib, and 13 samples were resistant to crizotinib. The drug resistance mechanisms in different samples are summarized in Figure 5A,B,C. In total, known resistance mutations were identified in 72 of the 117 patients who were resistant to first- or second-generation EGFR-TKIs, 22 of the 42 patients who were resistant to osimertinib, and 9 of the 13 patients who were resistant to crizotinib, which was similar to previous studies (20-25). Briefly, EGFR T790M was detected in 48 of the 117 patients who were resistant to first- or second-generation EGFR-TKIs, which was similar to the findings of a recent report (22). EGFR C797S/G, L718Q/V, and G719C were detected in 11 of the 42 patients who were resistant to osimertinib. Non-EGFR resistance mechanisms were identified in 94 patients who were resistant to EGFR-TKIs, with PI3K-AKT-mTOR signaling-related genomic alterations occurring in 16 patients. KRAS/NRAS and BRAF activation was detected in 9 and 3 patients, respectively. Gain-of-function mutations in ERBB2 were detected in 5 patients. MET amplification was identified in 9 patients. Finally, four patients in our cohort were identified to have ALK or RET fusion. Remarkably, PE supernatant outperformed plasma in identifying mutations that confer resistance to first- and second-generation EGFR-TKIs (75.4% vs. 29.8%, P<0.001, Figure 5D).
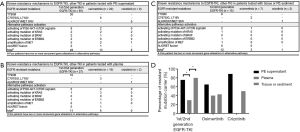
Discussion
This real-world investigation examined genetic alterations present in the supernatant of pleural effusion samples from individuals with metastatic NSCLC. The study recruited the largest cohort of individuals who had PE fluid for NGS testing to date. Real-world data demonstrated that analysis of cfDNA isolated from pleural effusion may provide important genomic information regarding an individual’s cancer that complements and expands data obtained from tissue biopsies.
Thoracentesis is widely used to relieve respiratory distress in patients with large amounts of PE. PE samples are collected from advanced cancer patients for therapeutic and diagnostic purposes. Moreover, due to its large quantity, PE can supply larger quantities of DNA for analyses compared with plasma. In our study, PE supernatant was used in almost half of NSCLC cases, which shows it to be the preferred choice for molecular profiling in the real-world setting (Figures 1,2).
The MSAFs of PE supernatant were significantly higher than that of plasma cfDNA in both cohort 1 and cohort 2, which corresponds with the findings other reports (16). This suggests that PE supernatant contains more abundant tumor-derived DNA. In cohort 1, more actionable mutations, such as ALK fusions and EGFR EX19 del mutation, were detected in supernatant samples compared with simultaneously collected samples of PE sediment or plasma. In cohort 2, the percentage of actionable EGFR, ALK, ROS1, RET, BRAF, KRAS, MET, and ERBB2 mutations was concordant with that of tissue reported in previous studies (19). However, plasma had significantly lower sensitivity in detecting EGFR actionable mutations in M1a disease but similar sensitivities in M1b/M1c disease (Figure 4C). Thus, for NSCLC patients with M1a stage disease, PE supernatant outperformed plasma for the detection of actionable mutations. We further explored the efficacy of PE supernatant in identifying resistance mutations. Similar to actionable mutations in treatment-naive patients, PE supernatant outperformed plasma for the detection of resistance mutations (Figure 5D). These data suggest that in a real-world setting, PE-supernatant offers a more accurate representation of tumor mutations than plasma in stage M1a patients when tumor tissues are not available.
The nature of PE formation has not been fully elucidated, although it is believed to originate in plasma, (26,27), the reasons for cfDNA being more highly concentrated in PE are still unknown. In advanced lung cancer, various mutation types, including single nucleotide variants (SNVs), InDels, fusions, and copy number variations, can be detected in PE supernatants. Several recent studies have shown PE supernatant to be more informative for tumor genomic profiling than plasma cfDNA. In our study, this was not only true for the initial diagnostic detection of sensitive mutations, but also for the identification of secondary mutations in drug-resistant tumors.
This study has some limitations. First, the cohort 1 had a small sample size, and no tissues samples were included. Therefore, we could not investigate the concordance between tissue DNA, plasma cfDNA, and PE supernatant and sediment at the same time point. Second, cohort 2 was a retrospective snapshot study, and no therapeutic efficacy data was considered.
Conclusions
This large-scale, real-world study verified that PE supernatant collected from advanced NSCLC patients had great reliability and efficacy in detecting mutations. For patients at with M1a stage disease, PE-supernatant is a better diagnostic tool and could serve as an alternative to tumor tissues. In clinical practice, PE supernatant is widely accepted by physicians not only for treatment naïve patients but also for treatment resistant patients.
Acknowledgments
Funding: This work was supported by the National Nature Science Foundation (81972188), Wu Jieping Medical Foundation (No. 320.6750.18125), State Key Laboratory of Respiratory Disease-The Open Project (SKLRD-OP-20180110), Beijing Municipal Science & Technology Commission (Z171100000417029), Sun Yat-sen University Young Teacher Plan (19ykpy179), and Guangzhou Science and Technology Program (202002020074).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-882
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-882
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-882). RC and YC are employees of Geneplus-Beijing Ltd. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the principles of the Declaration of Helsinki (as revised in 2013). The ethics committee of the First Affiliated Hospital of Nanjing Medical University (2019-SRFA-226) approved the study, and written informed consent was obtained from all patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Yang JC, Wu YL, Schuler M, et al. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol 2015;16:141-51. [Crossref] [PubMed]
- Gregorc V, Lazzari C, Karachaliou N, et al. Osimertinib in untreated epidermal growth factor receptor (EGFR)-mutated advanced non-small cell lung cancer. Transl Lung Cancer Res 2018;7:S165-70. [Crossref] [PubMed]
- Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472-84. [Crossref] [PubMed]
- Santarpia M, Liguori A, D’Aveni A, et al. Liquid biopsy for lung cancer early detection. J Thorac Dis 2018;10:S882-97. [Crossref] [PubMed]
- Haber DA, Velculescu VE. Blood-based analyses of cancer: circulating tumor cells and circulating tumor DNA. Cancer Discov 2014;4:650-61. [Crossref] [PubMed]
- Chen HF, Lei L, Wu LX, et al. Effect of icotinib on advanced lung adenocarcinoma patients with sensitive EGFR mutation detected in ctDNA by ddPCR. Transl Cancer Res 2019;8:2858-63. [Crossref]
- Stathopoulos GT, Kalomenidis I. Malignant pleural effusion: tumor-host interactions unleashed. Am J Respir Crit Care Med 2012;186:487-92. [Crossref] [PubMed]
- Benlloch S, Marti-Ciriquian JL, Galbis-Caravajal JM, et al. Cell-free DNA concentration in pleural fluid and serum: quantitative approach and potential prognostic factor in patients with cancer and pleural effusions. Clin Lung Cancer 2006;8:140-5. [Crossref] [PubMed]
- Kimura H, Fujiwara Y, Sone T, et al. High sensitivity detection of epidermal growth factor receptor mutations in the pleural effusion of non-small cell lung cancer patients. Cancer Sci 2006;97:642-8. [Crossref] [PubMed]
- Kimura H, Fujiwara Y, Sone T, et al. EGFR mutation status in tumour-derived DNA from pleural effusion fluid is a practical basis for predicting the response to gefitinib. Br J Cancer 2006;95:1390-5. [Crossref] [PubMed]
- Soh J, Toyooka S, Aoe K, et al. Usefulness of EGFR mutation screening in pleural fluid to predict the clinical outcome of gefitinib treated patients with lung cancer. Int J Cancer 2006;119:2353-8. [Crossref] [PubMed]
- Zhang X, Zhao Y, Wang M, et al. Detection and comparison of epidermal growth factor receptor mutations in cells and fluid of malignant pleural effusion in non-small cell lung cancer. Lung Cancer 2008;60:175-82. [Crossref] [PubMed]
- Gu J, Zang W, Liu B, et al. Evaluation of digital PCR for detecting low-level EGFR mutations in advanced lung adenocarcinoma patients: a cross-platform comparison study. Oncotarget 2017;8:67810-20. [Crossref] [PubMed]
- Li X, Liu Y, Shi W, et al. Droplet digital PCR improved the EGFR mutation diagnosis with pleural fluid samples in non-small-cell lung cancer patients. Clin Chim Acta 2017;471:177-84. [Crossref] [PubMed]
- Zhang P, Wu X, Tang M, et al. Detection of EGFR gene mutation status from pleural effusions and other body fluid specimens in patients with lung adenocarcinoma. Thorac Cancer 2019;10:2218-24. [Crossref] [PubMed]
- Tong L, Ding N, Tong X, et al. Tumor-derived DNA from pleural effusion supernatant as a promising alternative to tumor tissue in genomic profiling of advanced lung cancer. Theranostics 2019;9:5532-41. [Crossref] [PubMed]
- Zhuo M, Guan Y, Yang X, et al. The prognostic and therapeutic role of genomic subtyping by sequencing tumor or cell-free DNA in pulmonary large-cell neuroendocrine carcinoma. Clin Cancer Res 2020;26:892-901. [PubMed]
- Sun S, Liu Y, Eisfeld AK, et al. Identification of Germline Mismatch Repair Gene Mutations in Lung Cancer Patients With Paired Tumor-Normal Next Generation Sequencing: A Retrospective Study. Front Oncol 2019;9:550. [Crossref] [PubMed]
- Wang R, Zhang Y, Pan Y, et al. Comprehensive investigation of oncogenic driver mutations in Chinese non-small cell lung cancer patients. Oncotarget 2015;6:34300-8. [Crossref] [PubMed]
- Westover D, Zugazagoitia J, Cho BC, et al. Mechanisms of acquired resistance to first- and second-generation EGFR tyrosine kinase inhibitors. Ann Oncol 2018;29:i10-9. [Crossref] [PubMed]
- Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. [Crossref] [PubMed]
- Oxnard GR, Hu Y, Mileham KF, et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients With EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol 2018;4:1527-34. [Crossref] [PubMed]
- Le X, Puri S, Negrao MV, et al. Landscape of EGFR-Dependent and -Independent Resistance Mechanisms to Osimertinib and Continuation Therapy Beyond Progression in EGFR-Mutant NSCLC. Clin Cancer Res 2018;24:6195-203. [Crossref] [PubMed]
- Zhao J, Lin G, Zhuo M, et al. Next-generation sequencing based mutation profiling reveals heterogeneity of clinical response and resistance to osimertinib. Lung Cancer 2020;141:114-8. [Crossref] [PubMed]
- Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6:1118-33. [Crossref] [PubMed]
- Yano S, Shinohara H, Herbst RS, et al. Production of experimental malignant pleural effusions is dependent on invasion of the pleura and expression of vascular endothelial growth factor/vascular permeability factor by human lung cancer cells. Am J Pathol 2000;157:1893-903. [Crossref] [PubMed]
- Light RW, Hamm H. Malignant pleural effusion: would the real cause please stand up? Eur Respir J 1997;10:1701-2. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)