
Simultaneous analysis of ALK, RET, and ROS1 gene fusions by NanoString in Brazilian lung adenocarcinoma patients
Introduction
Lung cancer is the leading cause of cancer deaths in the world (1). Non-small cell lung cancer (NSCLC) accounts for the majority of lung cancer cases (85%), with adenocarcinoma being the most common histologic subtype (1,2). Most of the NSCLC cases are diagnosed in late stages when treatment has no curative intent. The development of oncogene-driven therapies has revolutionized the treatment of NSCLC dramatically, increasing the overall survival of advanced NSCLC patients (3-6). EGFR mutations are the most frequent actionable alterations in patients with lung adenocarcinomas (2,7,8). Other driver genomic alterations are ALK, RET, and ROS1 rearrangements, which can also be targeted for tyrosine kinase inhibitors (TKIs) (2,9). Although these genetic rearrangements are actionable, and the frequency of them is broadly investigated worldwide, the frequency of RET and ROS1 rearrangements is unknown in Brazilian patients (10-12). Moreover, Brazilian patients have a high admixture background, with contribution from European, African, Native American (Amerindian), and, more recently, Asian ethnicities (8,13,14). Therefore, the impact of actionable alterations on clinicopathological characteristics of NSCLC patients should be deeply explored (8,10,11,15,16).
Although significant efforts have been made on molecular techniques for the detection of actionable alterations in NSCLC, the scarcity of tumor cells in tumor biopsies due to the sampling procedures remains a challenge for molecular analysis. Targeted panels and multiplexed assays have been employed for optimizing molecular analysis of actionable genes in NSCLC, including a sequential approach for analysis’ feasibility on small tissue samples (17-22). We have previously reported the frequency of EGFR in 444 Brazilian lung adenocarcinomas and the association with Asian ancestry as well as the rate of KRAS mutations and its association with an unfavorable prognosis (8). In the present study, the ALK, RET, and ROS1 rearrangements were analyzed employing a single multiplex assay, and their associations with clinicopathological features and genetic ancestry were investigated. The detection of ALK, RET, and ROS1 rearrangements has a direct impact on clinical management of lung adenocarcinoma patients accruing in a shorter turnaround time through the employment of a single multiplexed assay enabling the tailored treatment as early as possible.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-740).
Methods
Study population and design
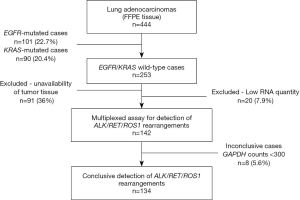
This retrospective study was conducted at the Molecular Oncology Research Center, the Department of Pathology, and the Department of Molecular Diagnosis, from patients diagnosed with lung adenocarcinoma (n=444) at Barretos Cancer Hospital from 2011 to 2014. All sociodemographics and clinicopathological data were collected retrospectively from medical records. Patients’ outcomes were collected from SISOnco (institutional software) and an active search was conducted when outcomes were not available from medical records (last updated: October 2019). Data on the main clinicopathological features and EGFR/KRAS mutation status and genetic ancestry of these cases were recently reported (8). Since the main molecular alterations in lung adenocarcinoma are well known to be mutually exclusive with EGFR and KRAS mutations, only EGFR / KRAS wild-type lung adenocarcinoma cases (n=253) were enrolled in the present study (Figure 1). However, due to a lack of available tumor tissue (n=91) or low RNA quantity (lower than 100 ng, n=20), gene fusions were evaluated in 142 EGFR/KRAS wild-type cases (Figure 1).
This study was approved by the local ethics committee (Barretos Cancer Hospital IRB/Project No. 630/2012), with the exemption of informed consent. The methodology was performed following the Declaration of Helsinki (as revised in 2013).
RNA isolation
RNA isolation was performed from formalin-fixed paraffin-embedded (FFPE) tumor samples, sectioned on slides with a thickness of 10µm as previously reported (23). One slide was stained with hematoxylin and eosin (H&E) and evaluated by an experienced pathologist for identification, sample adequacy assessment, and selection of the tumor tissue area (minimum of 70% tumor area). RNA was isolated using a commercial kit (RNeasy FFPE Mini Kit, Qiagen, Hilden, Germany) according to the manufacturer's instructions.
Simultaneous detection of ALK, RET and ROS1 rearrangements by NanoString custom panel
Detection of ALK, RET, and ROS1 rearrangements was performed in 142 out of the 253 EGFR/KRAS wild-type cases since, in 91 cases, the tissue was unavailable and in 20 cases in which the RNA quantity was insufficient (Figure 1). The nCounter® Elements XT (NanoString Technologies, Seattle, WA, USA) custom panel was designed following previously described techniques for the evaluation of transcripts using specific probes. It contained 24 probes for the 5' and 3' regions of the ALK, RET and ROS1 genes along with 12 specific probes for the rearrangement partners (EML4-ALK, KIF5B-ALK, TGF-ALK; CCDC6-RET, KIF5B-RET; CD74-ROS1, EZR-ROS1, GOPC-ROS1, LRIG3-ROS1, SLC34A2-ROS1, TPM3-ROS1, SDC4-ROS1) (20). Briefly, from 100 to 300 ng of total RNA samples were hybridized with specific probes for 21 hours at 67 °C. The hybridized complexes were purified in the PrepStation (NanoString Technologies) and immobilized in the cartridge. The cartridge was scanned by the Digital Analyzer (NanoString Technologies) for counting the transcripts (24). The positive controls used were an ALK-positive cell line (H2228 cell line) and a commercial control harboring ALK, RET, and ROS1 rearrangements (Horizon Discovery, Cat. No.: HD784).
The transcripts counts were normalized by the nSolver Analysis® Software v4.0 (NanoString Technologies), using the ratio of geometric mean for each sample and arithmetic mean of all samples for positive assay controls and reference genes (housekeepings). Inconclusive results were considered when counts lower than 300 counts were obtained for GAPDH. The calculation of the imbalance probes was defined by the ratio between geometric mean of 3' probes and the average of 5' probes, considering thresholds for ALK rearrangement positivity equal to 2, for RET equal to 5 and for ROS1 equal to 3, as previously reported (20,22). Imbalance analysis, detection of fusion partners (count more of 50), and graphical construction was performed in R environment v3.4.1 with scripts implemented in the local Galaxy server (25).
Detection of ALK, RET and ROS1 rearrangements by NanoString Lung Fusion Panel (nCounter Vantage 3D™)
Validation of the presence of ALK, RET, and ROS1 rearrangements was performed using the Lung Fusion assay (NanoString Technologies). This assay was designed for the evaluation of transcripts using specific probes for the 5' and 3' regions from ALK, RET, and ROS1 genes and specific probes for the rearrangement partners from ALK, RET, ROS1 and NTRK1 (no NTRK imbalance probes are provided in this assay). The transcription counts were normalized by the nSolver Analysis® Software v4.0 (NanoString Technologies). The calculation of the imbalance between the 3' and 5' probes was performed by a t-test comparing the log-scale data from probes. A significant P value provided the final positive result about fusion presence from the t-test plus the detection of fusion partners. These analyses were conducted using the Advanced Analysis v2.0 package (NanoString Technologies).
Fluorescence in situ hybridization (FISH)
Detection of RET and ROS1 rearrangements was performed using commercial probes (ZytoLight SPEC RET Dual Color Break Apart, ZytoLight SPEC ROS1 Dual Color Break Apart). Breast adenocarcinoma tissue was used as a negative control. For considering the sample suitable for evaluation, more than 15% of positive cells in at least 100 cells should be present. Hybridization reactions were repeated twice. FISHView 7.0 software (Applied Spectral Imaging) was employed for the analysis.
Ancestry analysis
The genetic ancestry background was previously assessed in the tumor DNA by a panel of 46-ancestry informative markers that allow estimating the ancestral proportions of African (AFR), European (EUR), Asian (ASN), and Native American (AME) populations (8).
Statistical analysis
Univariate (t-test/ Fisher’s exact test/ chi-square test) and multivariate (Linear regression model) analyses were used to determine if gene rearrangements had a significant effect on the investigated parameters. Kaplan-Meier method and Log-rank test were used for univariate survival analysis, and Cox, the proportional hazards model, was used for multivariate survival analysis. For survival analysis, death was considered as an event, and live patients or patients who lost follow up were considered as censored. The survival analysis was conducted only for patients diagnosed at stage IV to decrease bias regarding the clinical outcome. Statistical analysis performed by IBM SPSS® Statistics Base software (IBM, Armonk, NY) with a 95% significance limit.
Results
Clinicopathological and sociodemographic features
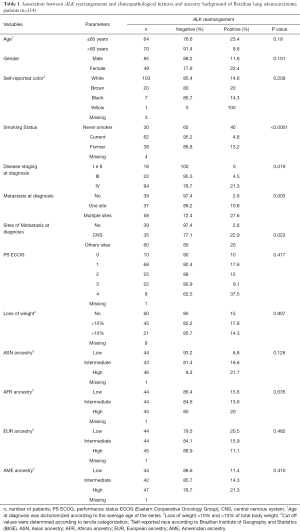
The NanoString results were conclusive in 94.4% of the cases (n=134), resulting in a failure rate of 5.6% (8 out of 142 presented inconclusive results) (Figure 1). Overall, the majority of the patients was male (63%; n=85), with an average age of 60 years old, self-reported as white (77%; n=103), current or former smokers (75%; n=62 and n=38, respectively), diagnosed at stage IV (70%; n=94) and presented with metastasis in multiple sites at diagnosis (43%; n=58) (Table 1).
Full table
ALK rearrangements and clinicopathological associations
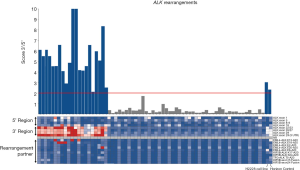
ALK rearrangements were detected in 15.7% of the EGFR/KRAS wild-type cases (21 out of 134) (Figure 2) (Table S1). The most frequently observed rearrangement partner was EML4-ALK (33.3%; 7 out of 21) (Figure 2). As expected, both commercial control and H2228 cell line were positive for ALK rearrangement, validating the assay. When considering all the 325 molecularly analyzed cases (134 plus 101 EGFR-mutated and 90 KRAS-mutated cases), we observed a frequency of 6,5% (21/325) of ALK gene fusions.
The presence of ALK rearrangements was associated with younger age at diagnosis (P=0.049), never smokers (P<0.0001), disease stage IV at diagnosis (P=0.019), metastases in multiple sites (P=0.003) and presence of metastases in central nervous system (CNS) (P=0.023; Table 1). Gender, self-reported color, loss of weight, genetic ancestry, ECOG PS were not associated with ALK rearrangements. The presence of ALK rearrangements was not associated with clinical outcome (P=0.486; Figure S1).
In multivariate analysis, the presence of ALK rearrangements was associated with never smokers (OR =12.432; P<0.0001; Table 2) and presence of metastasis in central nervous system (CNS) (OR =13.224; P=0.029; Table 2).

Full table
RET and ROS1 rearrangements
RET and ROS1 rearrangements were detected in two and one out of 134 cases, respectively (Figures 3,4) (Table S1). No rearrangement partner was identified in the RET and ROS1 positive cases. No statistical associations with the clinicopathological characteristics could be performed due to the low sample size. Overall, RET positive cases were male, never smokers, diagnosed at stage IV, and presented with metastases in the CNS. ROS1 positive case was female, never smoker, diagnosed at stage IV, and presented with lymph node metastases.
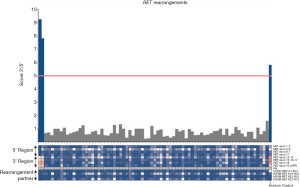
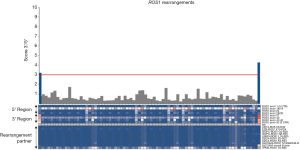
When considering all the 325 molecularly for which a molecular analysis was conclusive (134 plus 101 EGFR-mutated and 90 KRAS-mutated cases), we observed a frequency of 0.6% (2/325), and 0.3% (1/325), for RET and ROS1, respectively.
Validation of the ALK, RET and ROS1 rearrangements
ALK rearrangements detection by NanoString using the same gene panel was previously validated by immunohistochemistry by our group (23). Concerning RET and ROS1, rearrangements were further analyzed by FISH; however the experiments were considered inconclusive due to the insufficient number of signals observed, probably due to pre-analytical issues of the tissue (Figure S2).
Due to unsuccessful attempts for confirming RET and ROS1 rearrangements by FISH, we further validated the results for the cases exhibiting RET (n=2) and ROS1 rearrangements (n=1) with the commercial Lung Fusion assay (Figure S3). Both RET-positive samples were confirmed as positive by the Lung Fusion assay (Figure S3A and S3C). In one of then, the partner was identified-KIF5B (Figure S3F). Although the known KIF5B-RET fusion partner was included in our custom panel, the variant (variant 12) that was detected is not included in our custom panel.
The ROS1-positive sample was also confirmed as positive by the Lung Fusion assay (Figure S3B), and no fusion partner was identified (Figure S3E).
Ancestry analysis
The mean of ancestry proportions observed among the 134 NSCLC patients was 74.7% for the EUR, 13.1% for the AFR, 5.8% for the AME, and 6.4% for the ASN (Figure S4). The mean of ancestry proportions observed among the 444 NSCLC patients was 73.1% for the EUR, 13.1% for the AFR, 6.5% for the AME, and 7.3% for the ASN (Figure S5 and Table S2) as previously reported (8). In accordance with a high percentage of EUR, most patients were self-declared white (Table 1). The presence of ALK rearrangements was not correlated with genetic ancestry (Table 1).
Although Barretos Cancer Hospital is located in the upstate of Sao Paulo, it is a reference center that assists patients from all over the country. Although not all Brazilian states are currently represented in our series and the Southeast of Brazil is the most represented region, all ancestry proportions are represented in the current series (Figure S6).
Discussion
The identification of actionable molecular alterations has conferred therapeutic relevance for advanced NSCLC patients. Although several studies have reported the frequency of these molecular alterations, they were performed mostly in European and Asian populations. Data on admixture populations remain lacking. In this study, we reported the frequency of ALK, RET, ROS1 rearrangements in a Brazilian series of lung adenocarcinoma using a single multiplexed assay, and the association of these rearrangements with clinicopathological features and ancestry.
In the present series, we have previously reported the frequency of EGFR (22.7%) and KRAS mutations (20.4%) (8). Regarding ALK, RET, and ROS1 rearrangements, it is well known that they are all mutually exclusive and mutually exclusive with other driver mutations, so only EGFR and KRAS wild-type cases were evaluated (26-30).
We identified ALK rearrangements in 6.5% (21/325) of lung adenocarcinomas, which is in line with the reported worldwide. The frequency of ALK rearrangements in the literature varies from 3% to 10.8% (31-33). In Latin America, a study enrolling 5,130 NSCLC patients from 10 countries (excepting Brazil), the frequencies of ALK rearrangements ranged from 4.1% to 10.8% (Colombia 4.1%; Panama 4.4%; Uruguay 5.4%; Argentina 6.0%; Mexico 7.6%; Chile 8.6%; Venezuela 8.9%; Costa Rica 9.5%; Peru 10.8%) (34). In Brazil, two recent studies employing immunohistochemistry, one from Northeast and another from South, reported frequencies of ALK rearrangements of 10.4% (n=173 patients) and 4% (n=275 patients), respectively (10,12). The frequency of ALK rearrangements in the present work is in between other Brazilian studies. Such variation could be explained by the admixture background of the Brazilian population (35), since ancestry background is a factor that may influence the frequency of actionable alterations (36). Although the Southeast of Brazil is the most represented region in the current series, all ancestry proportions are indeed represented. Yet, our ancestry analysis did not identify any significant association.
We observed that the presence of the ALK rearrangements was associated with younger age, never smokers, and the presence of metastases in CNS, in line with some studies (37,38). Considering that ALK-positive patients are recurrently younger and associated with advanced disease at diagnosis, these cases may be more aggressive even from the beginning of the disease. For this reason, the identification of driver alterations becomes even more important to guide treatment with targeted therapies. ALK-positive patients are eligible for treatment with crizotinib (5). In this present study, only one ALK-positive patient enrolled in a clinical trial (phase III-2013) was treated with crizotinib. Although treatment with ALK inhibitors is approved by the Brazilian regulatory agency (ANVISA), patients do not have access to treatment via the public health system (39).
Regarding RET and ROS1 rearrangements, the frequency observed in our overall series was 0.6% (2/325) and 0.3% (1/325), respectively. The frequency of RET rearrangements in lung adenocarcinoma is lower than 2% worldwide, ranging from 0.2-1.9% in Asian patients and 1.3% in patients from USA (27,32,40-43). The frequency of ROS1 rearrangements in lung adenocarcinoma is variable, ranging from 1.2% in Europeans patients and reaching 3.4% in Chinese patients (40,42,44,45). There are no reports on the presence of RET and ROS1 rearrangements in the Brazilian or any other admixture population. Due to the small number of RET and ROS1-positive cases, no statistical analysis could be performed. Anyhow, similarly to ALK-positive cases, the presence of RET and ROS1 rearrangements were observed in younger patients, and never smokers (36,40,42,43,46,47). Despite the shallow frequency of RET and ROS1 rearrangements, they constitute important therapeutic actionable alterations. The ALK-inhibitors, have also demonstrated success in the treatment of ROS1-positive patients, and it was approved by USA-FDA, and Brazilian ANVISA for these patients (4,48). Concerning RET rearrangements, the TKi selpercatinib (LOXO-292) has recently been approved by the FDA for the treatment of lung and thyroid tumors, due to the highly promising results obtained by clinical trials (ClinicalTrials.gov: NCT04268550 and ClinicalTrials.gov: NCT03157128) (49-51).
Due to a large number of driver alterations and the scarcity of tumor tissue usually available for molecular testing of NSCLC patients, the use of multiplexed assay platforms for FFPE can be a powerful tool. The NanoString technology is very robust, sensitive, easy to execute, with multiplex capabilities, and more cost-effective when custom panels are employed, surpassing FISH, IHC, and NGS techniques (22,23). We have previously shown that the NanoString platform managed to identify the presence of ALK rearrangements in FFPE samples, highlighting its capacity to detect RNA transcripts in highly degraded samples employing low RNA input (23). Likewise, in the extended study, the detection of rearrangements in FFPE samples was feasible, even in samples that lead to inconclusive FISH results. Moreover, due to the multiplex capability of the NanoString platform, it was possible to detect both gene rearrangements and known partners (20,22,23). Our panel includes the most observed rearrangement partners for each gene investigated as well as variants of these partners. Thus, it was possible to identify the rearrangement partners for ALK and RET-positive cases in our study. Importantly, the possibility of custom panels, allowed to add novel gene rearrangements and new variants from fusion partners, such as with NTRK rearrangements (18,52). This maximizes the number of patients for oncogene-driven therapy for NSCLC with no additional tissue sample required and minimally increased in the test cost. In addition, the employment of a single multiplexed assay accrues in a shorter turnaround time for report release enabling the tailored treatment as early as possible.
One limitation of the present study is that the number of patients in the present study does not portrait the entire Brazilian lung cancer population, and further studies analyzing a higher number of cases and patients from all regions of Brazil are warranted.
Conclusions
Our study successfully used a single assay for the detection of ALK, RET, and ROS1 fusions in FFPE biopsies of lung adenocarcinoma. The NanoString methodology allows an extensive molecular investigation of the significant actionable gene rearrangements potentially to be employed in the diagnostic routine for contributing to better guide clinical treatment strategies for Brazilian patients with lung adenocarcinoma.
Acknowledgments
We thank Barretos Cancer Hospital for supporting the present study and Center of Molecular Diagnosis and Molecular Oncology Research Center (Barretos Cancer Hospital, Barretos, Brazil) for provide proper infrastructure. We thank Adeylson Ribeiro for epidemiological and geographical support, including the design of Figure S6. We also thank GTOP (Translacional Group of Pulmonary Oncology) for all discussions.
Funding: The research was supported by Barretos Cancer Hospital, Barretos, Brazil; FINEP (MCTI/FINEP/MS/SCTIE/DECIT - BioPlat 1302/13); and Public Ministry of Labor Campinas (Research, Prevention, and Education of Occupational Cancer) in Campinas, Brazil. LACN is supported by the National Ministry of Health (Brazil). LFL and LSS are supported by the Public Ministry of Labor Campinas (Research, Prevention, and Education of Occupational Cancer) in Campinas, Brazil. RMR is a recipient of a CNPq Productivity (Brazil) fellowship.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-740
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-740
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-740). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the local ethics committee (Barretos Cancer Hospital IRB/ Project No. 630/2012), with the exemption of informed consent. The methodology was performed following the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Testa U, Castelli G, Pelosi E. Lung Cancers: Molecular Characterization, Clonal Heterogeneity and Evolution, and Cancer Stem Cells. Cancers (Basel) 2018;10:248. [Crossref] [PubMed]
- Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [Crossref] [PubMed]
- Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014;371:1963-71. [Crossref] [PubMed]
- Solomon BJ, Mok T, Kim DW, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med 2014;371:2167-77. [Crossref] [PubMed]
- Yun PJ, Wang GC, Chen YY, et al. Brain metastases in resected non-small cell lung cancer: The impact of different tyrosine kinase inhibitors. PLoS One 2019;14:e0215923. [Crossref] [PubMed]
- Shtivelman E, Hensing T, Simon GR, et al. Molecular pathways and therapeutic targets in lung cancer. Oncotarget 2014;5:1392-433. [Crossref] [PubMed]
- Leal LF, de Paula FE, De Marchi P, et al. Mutational profile of Brazilian lung adenocarcinoma unveils association of EGFR mutations with high Asian ancestry and independent prognostic role of KRAS mutations. Sci Rep 2019;9:3209. [Crossref] [PubMed]
- Oberndorfer F, Mullauer L. Molecular pathology of lung cancer: current status and perspectives. Curr Opin Oncol 2018;30:69-76. [Crossref] [PubMed]
- Andreis TF, Correa BS, Vianna FS, et al. Analysis of Predictive Biomarkers in Patients With Lung Adenocarcinoma From Southern Brazil Reveals a Distinct Profile From Other Regions of the Country. J Glob Oncol 2019;5:1-9. [Crossref] [PubMed]
- Machado-Rugolo J, Fabro AT, Ascheri D, et al. Usefulness of complementary next-generation sequencing and quantitative immunohistochemistry panels for predicting brain metastases and selecting treatment outcomes of non-small cell lung cancer. Hum Pathol 2019;83:177-91. [Crossref] [PubMed]
- Oliveira AC, Silva A, Alves M, et al. Molecular profile of non-small cell lung cancer in northeastern Brazil. J Bras Pneumol 2019;45:e20180181. [Crossref] [PubMed]
- Adhikari K, Chacon-Duque JC, Mendoza-Revilla J, et al. The Genetic Diversity of the Americas. Annu Rev Genomics Hum Genet 2017;18:277-96. [Crossref] [PubMed]
- Moura RR, Coelho AV, Balbino Vde Q, et al. Meta-analysis of Brazilian genetic admixture and comparison with other Latin America countries. Am J Hum Biol 2015;27:674-80. [Crossref] [PubMed]
- Bacchi CE, Ciol H, Queiroga EM, et al. Epidermal growth factor receptor and KRAS mutations in Brazilian lung cancer patients. Clinics 2012;67:419-24. [Crossref] [PubMed]
- Carneiro JG, Couto PG, Bastos-Rodrigues L, et al. Spectrum of somatic EGFR, KRAS, BRAF, PTEN mutations and TTF-1 expression in Brazilian lung cancer patients. Genet Res (Camb) 2014;96:e002. [Crossref] [PubMed]
- Cohen D, Hondelink LM, Solleveld-Westerink N, et al. Optimizing Mutation and Fusion Detection in NSCLC by Sequential DNA and RNA Sequencing. J Thorac Oncol 2020;15:1000-14. [Crossref] [PubMed]
- Karlsson A, Cirenajwis H, Ericson-Lindquist K, et al. A combined gene expression tool for parallel histological prediction and gene fusion detection in non-small cell lung cancer. Sci Rep 2019;9:5207. [Crossref] [PubMed]
- Lindquist KE, Karlsson A, Leveen P, et al. Clinical framework for next generation sequencing based analysis of treatment predictive mutations and multiplexed gene fusion detection in non-small cell lung cancer. Oncotarget 2017;8:34796-810. [Crossref] [PubMed]
- Lira ME, Choi YL, Lim SM, et al. A single-tube multiplexed assay for detecting ALK, ROS1, and RET fusions in lung cancer. J Mol Diagn 2014;16:229-43. [Crossref] [PubMed]
- Reguart N, Teixido C, Gimenez-Capitan A, et al. Identification of ALK, ROS1, and RET Fusions by a Multiplexed mRNA-Based Assay in Formalin-Fixed, Paraffin-Embedded Samples from Advanced Non-Small-Cell Lung Cancer Patients. Clin Chem 2017;63:751-60. [Crossref] [PubMed]
- Rogers TM, Arnau GM, Ryland GL, et al. Multiplexed transcriptome analysis to detect ALK, ROS1 and RET rearrangements in lung cancer. Sci Rep 2017;7:42259. [Crossref] [PubMed]
- Evangelista AF, Zanon MF, Carloni AC, de Paula FE, Morini MA, Ferreira-Neto M, et al. Detection of ALK fusion transcripts in FFPE lung cancer samples by NanoString technology. BMC Pulm Med 2017;17:86. [Crossref] [PubMed]
- Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 2008;26:317-25. [Crossref] [PubMed]
- Afgan E, Baker D, Batut B, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 2018;46:W537-44. [Crossref] [PubMed]
- Won JK, Keam B, Koh J, et al. Concomitant ALK translocation and EGFR mutation in lung cancer: a comparison of direct sequencing and sensitive assays and the impact on responsiveness to tyrosine kinase inhibitor. Ann Oncol 2015;26:348-54. [Crossref] [PubMed]
- Dugay F, Llamas-Gutierrez F, Gournay M, et al. Clinicopathological characteristics of ROS1- and RET-rearranged NSCLC in caucasian patients: Data from a cohort of 713 non-squamous NSCLC lacking KRAS/EGFR/HER2/BRAF/PIK3CA/ALK alterations. Oncotarget 2017;8:53336-51. [Crossref] [PubMed]
- Seto K, Kuroda H, Yoshida T, et al. Higher frequency of occult lymph node metastasis in clinical N0 pulmonary adenocarcinoma with ALK rearrangement. Cancer Manag Res 2018;10:2117-24. [Crossref] [PubMed]
- Tang Z, Zhang J, Lu X, et al. Coexistent genetic alterations involving ALK, RET, ROS1 or MET in 15 cases of lung adenocarcinoma. Mod Pathol 2018;31:307-12. [Crossref] [PubMed]
- Gainor JF, Varghese AM, Ou SH, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res 2013;19:4273-81. [Crossref] [PubMed]
- Liu L, Liu J, Shao D, Deng Q, Tang H, Liu Z, et al. Comprehensive genomic profiling of lung cancer using a validated panel to explore therapeutic targets in East Asian patients. Cancer Sci 2017;108:2487-94. [Crossref] [PubMed]
- Levy MA, Lovly CM, Pao W. Translating genomic information into clinical medicine: lung cancer as a paradigm. Genome Res 2012;22:2101-8. [Crossref] [PubMed]
- Ross JS, Ali SM, Fasan O, Block J, Pal S, Elvin JA, et al. ALK Fusions in a Wide Variety of Tumor Types Respond to Anti-ALK Targeted Therapy. Oncologist 2017;22:1444-50. [Crossref] [PubMed]
- Arrieta O, Cardona AF, Bramuglia G, et al. Molecular Epidemiology of ALK Rearrangements in Advanced Lung Adenocarcinoma in Latin America. Oncology 2019;96:207-16. [Crossref] [PubMed]
- Souza AM, Resende SS, Sousa TN, et al. A systematic scoping review of the genetic ancestry of the Brazilian population. Genet Mol Biol 2019;42:495-508. [Crossref] [PubMed]
- Calvayrac O, Pradines A, Pons E, et al. Molecular biomarkers for lung adenocarcinoma. Eur Respir J 2017;49:1601734. [Crossref] [PubMed]
- Fan L, Feng Y, Wan H, et al. Clinicopathological and demographical characteristics of non-small cell lung cancer patients with ALK rearrangements: a systematic review and meta-analysis. PLoS One 2014;9:e100866. [Crossref] [PubMed]
- Yamaguchi N, Vanderlaan PA, Folch E, et al. Smoking status and self-reported race affect the frequency of clinically relevant oncogenic alterations in non-small-cell lung cancers at a United States-based academic medical practice. Lung Cancer 2013;82:31-7. [Crossref] [PubMed]
- Araujo LH, Baldotto C, Castro G Jr, et al. Lung cancer in Brazil. J Bras Pneumol 2018;44:55-64. [Crossref] [PubMed]
- Davies KD, Le AT, Theodoro MF, et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin Cancer Res 2012;18:4570-9. [Crossref] [PubMed]
- Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med 2012;18:375-7. [Crossref] [PubMed]
- Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med 2012;18:378-81. [Crossref] [PubMed]
- Wang R, Hu H, Pan Y, et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol 2012;30:4352-9. [Crossref] [PubMed]
- Fu S, Liang Y, Lin YB, et al. The Frequency and Clinical Implication of ROS1 and RET Rearrangements in Resected Stage IIIA-N2 Non-Small Cell Lung Cancer Patients. PLoS One 2015;10:e0124354. [Crossref] [PubMed]
- Zhong S, Zhang H, Bai D, et al. Detection of ALK, ROS1 and RET fusion genes in non-small cell lung cancer patients and its clinicopathologic correlation. Zhonghua Bing Li Xue Za Zhi 2015;44:639-43. [PubMed]
- Bergethon K, Shaw AT, Ou SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 2012;30:863-70. [Crossref] [PubMed]
- Warth A, Muley T, Dienemann H, et al. ROS1 expression and translocations in non-small-cell lung cancer: clinicopathological analysis of 1478 cases. Histopathology 2014;65:187-94. [Crossref] [PubMed]
- ANIVISA. Aprovação Crizotinib 2016 [cited 2018 december,27]. Available online: https://consultas.anvisa.gov.br/#/medicamentos/25351725846201208/
- Drilon A OG, Wirth L, et al. A phase 1/2 trial of LOXO-292 in patients with RET fusion-positive lung cancers. IASLC 2019 World Conference on Lung Cancer hosted by the International Association for the Study of Lung Cancer; September 9, 2019; Barcelona, Spain 2019.
- Guo R, Schreyer M, Chang JC, et al. Response to Selective RET Inhibition With LOXO-292 in a Patient With RET Fusion-Positive Lung Cancer With Leptomeningeal Metastases. JCO Precis Oncol 2019;3:PO.19.00021.
- Halliday PR, Blakely CM, Bivona TG. Emerging Targeted Therapies for the Treatment of Non-small Cell Lung Cancer. Curr Oncol Rep 2019;21:21. [Crossref] [PubMed]
- Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N Engl J Med 2018;378:731-9. [Crossref] [PubMed]


