
A prognostic nomogram for lung adenocarcinoma based on immune-infiltrating Treg-related genes: from bench to bedside
Introduction
Lung cancer is the leading cause of malignancy-related death worldwide, with approximately 2.2 million new cases and 1.9 million deaths worldwide in 2017 (1), despite great progress in diagnosis and therapeutics over the years. As the most prevailing histological type of lung cancer, lung adenocarcinoma (LUAD) always derives from the acinar cells of the lung periphery and has a poor prognosis. This cause may be mainly due to the high heterogeneity of LUAD and advanced stage of patients when diagnosed (2). However, the current TNM (tumor size, lymph nodes and distant metastasis) staging system tends to be insufficient to accurately characterize and stage tumors at an early stage and even after surgery (3,4), features that are necessary to design an optimal initial treatment plan and offer adequate postoperative adjuvant therapy. Therefore, more complementary methods to enhance TNM staging and identify the behavior of LUAD are needed.
In recent years, our view of cancer has changed drastically. Tumors are no longer regarded as simple malignant masses or cells but as a complex tumor microenvironment: tumor cells recruit other infiltrating immune cell subpopulations to constitute a self-sufficient biological unit (5). The composition of the tumor microenvironment varies in different patients and even in the same type of cancer, such as different fractions of B cells, NK cells, M1/M2 macrophages, granulocytes, mast cells, CD8+ T cells, CD4+ helper T cells, and regulatory T cells, which determine tumor characteristics and the patient prognosis (6-8). Previous studies showed that the expansion and accumulation of suppressive Tregs always caused the development, metastasis and recurrence of multiple malignancies including lung cancer (9-12). Some studies found that FoxP3+CD4+Tregs infiltrating correlated negatively with the survival of small cell lung cancer (SCLC) (13,14). Other researches on non-small cell lung cancer (NSCLC) revealed that whether in peripheral blood or intratumor, high level of Tregs was associated with high metastasis and low survival rates (15-17). However, a study published in ‘Nature Immunology’ from Ferreira et al., highlighted the role of type 1 Tregs in enhancing the immunity barrier in peripheral tissues, which challenged the classical view of Tregs in immunosuppression (18). Given that the role of Tregs (regulatory T cells) in tumors has been controversial, we attempted to explore the potential value of Tregs for LUAD.
In the present study, we integrated three LUAD sequencing datasets, GSE10072, GSE32863, and GSE43458, from the Gene Expression Omnibus (GEO) database and extracted the relevant fractions of 22 immune-infiltrating cells from the CIBERSORTx portal. Next, weighted gene coexpression network analysis (WGCNA) and protein-protein interaction (PPI) network analysis were conducted to identify the most significant module and candidate genes related to Tregs. The candidate genes were then further validated using data from The Cancer Genome Atlas (TCGA) database, and 13 hub genes were screened. The correlation between hub genes and Tregs was tested using Spearman’s method. Finally, we constructed a nomogram model combining the hub genes and clinical parameters, which showed a better performance to predict the risk of LUAD. The flow diagram of this study is shown in Figure 1. We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-822).
Methods
Data source and processing
The LUAD sequencing data were applied from three GEO (http://www.ncbi.nlm.nih.gov/geo/) datasets, GSE10072, GSE32863, and GSE43458. We used the sav and limma packages of R to perform batch calibration and data normalization. When a gene corresponds to multiple probes, the mean value is taken as the final expression value.
Estimation of immune infiltrating cells
Using the sequencing data, we estimated the fractions of 22 tumor-infiltrating immune cells using CIBERSORTx (https://cibersortx.stanford.edu/), an online tool that imputes gene expression profiles by a deconvolution algorithm and provides an estimated abundance of known cell types within a mixed cell population (19).
Construction of the coexpression network and module-trait relationships
The expression values of the 10,048 genes of the LUAD samples were used to construct a weight coexpression network employing the R package “WGCNA”, a biological method used to integrate genes with coexpression into the same module. The correlations between the modules and sample traits are calculated to screen the models with a high correlation with traits, and the genes in the modules are analyzed to identify target genes (20).
Here, we used the fractions of 22 immune-infiltrating cells as sample traits and chose an optimal soft threshold power (β) to build a scaleless network when setting the index of scale-free topologies as 0.90. Next, we assigned genes with similar expression patterns to the same module (minimum size =30) using the “dynamic tree cutting” algorithm. Moreover, we estimated the correlation of the module eigengenes with the infiltrating level of the 22 immune-infiltrating cells to screen the significance of the modules by Pearson’s test. Finally, we selected the “Tregs (regulatory T cells)” subtype of interest and the module with the highest correlation with Tregs was selected for further study.
Construction of the PPI network and identification of candidate genes
From the significant module, we obtained Treg-related module genes, with which the PPI network was developed using Search Tool for the Retrieval of Interacting Genes (STRING; https://string-db.org/). Next, the PPI network was presented using Cytoscape (version 3.7.2), which is a free app for visualizing sophisticated networks and integrating them with attribute data. The “CytoHubba” module of Cytoscape is a plug-in that recognizes hub genes in a network based on the properties of nodes in a network (21), from which we screened the candidate genes related to Tregs.
Verification of candidate genes using TCGA data and screening of the hub genes
To further verify the role of candidate genes in LUAD, we applied gene sequencing and the corresponding completely clinical data (375 LUAD and 48 normal samples) at the TCGA (https://portal.gdc.cancer.gov/) portal, from which we extracted the expression values of the Treg-related genes. The expression values of LUAD and normal tissues were averaged using the “mean” function. Next, we used log2 transform to normalize all the average expression values. Analysis of the statistically significant differences between the LUAD and normal expression data was conducted using the Wilcoxon signed-rank test built into R (version 3.6.3; https://www.r-project.org/), defining the threshold of |log(fold change)| no less than 1 and the false discovery rate (FDR) less than 0.05. Finally, we implemented a Cox proportional hazards model to screen the Treg-related hub genes and their coefficients, from which the patients were assigned a high- or low-risk score. The risk scores were calculated using the following formula:
Validation of Treg-related hub genes
The Kaplan-Meier (K-M) curve was illustrated to estimate the differences in overall survival (OS) between the low- and high-risk groups using the log-rank test to analyze the statistical significance. Moreover, we implemented receiver operating characteristic (ROC) curves to evaluate the accuracy of grouping (low/high risk). Spearman’s correlation between the Treg infiltration level and expression of hub genes was calculated using the data from Tumor Immune Estimation Resource (TIMER2.0; http://timer.cistrome.org/), and the results were visualized using the “ggstatsplot” package of R.
Enrichment analysis of hub genes related to Tregs
To identify tumor-related molecular mechanisms of the hub genes related to Tregs, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed in R using the packages org.Hs.eg.db, Cluster Profiler, enrichplot, and ggplot2 with both P values and q-values less than 0.05.
Construction and validation of the prognostic nomogram model for LUAD
To apply the Treg-related genes better clinically, we constructed a prognostic nomogram model for LUAD, combining the risk score with traditional clinical parameters (age, gender, stage, T, N and M). A nomogram is an effective tool that formulates the scoring criteria for all the variables in the regression equation according to their regression coefficients. Next, each patient receives a summed score, which can be converted into the probability of the outcome time of each patient through the function (22). We then performed ROC, calibration and K-M curve analyses to elucidate the performance of the nomogram.
Statistical analysis
All statistical analyses and graphics were generated using the R and Perl packages. A Cox proportional hazards model was applied to identify survival genes related to Tregs. K-M curve analysis was performed to show the differences in OS between the low- and high-risk groups, using the log-rank test to estimate the significance of the differences. The calibration curve, ROC curve and area under the curve (AUC) values were used to determine the efficacy of the model. A P value less than 0.05 was defined as statistically significant.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Gene sequencing data and estimation of immune-infiltrating cells
We acquired the gene expression data of 10,048 genes from 196 LUAD and 137 normal tissues from the GEO database and calculated the abundance of 22 immune-infiltrating cells for each sample using the CIBERSORTx portal. Next, the fractions of 22 immune-infiltrating cells with gene expression data were selected as traits of WGCNA.
Coexpression network and protein-protein interaction network
The expression data of the 10,048 genes with the abundance of 22 immune-infiltrating cells were used to build the coexpression network. To build the scaleless network, we chose the appropriate soft threshold power (β=10) because it was the first power value to make the index of scale-free topologies reach 0.90 (Figure 2A,B). Treg-related genes with similar expression patterns were incorporated into the same module using a dynamic tree-cutting algorithm (module size =30), making a hierarchical clustering tree with 12 modules (Figure 2C). As shown in Figure 3A, the black module was highly correlated with regulatory T cells (Tregs) (R2=0.52, P=1e-10) among the twelve modules. Because we were interested in Tregs, we selected 111 Treg-related genes (Table S1) in the black module with P<0.05 for further study.
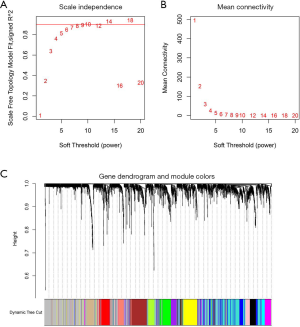
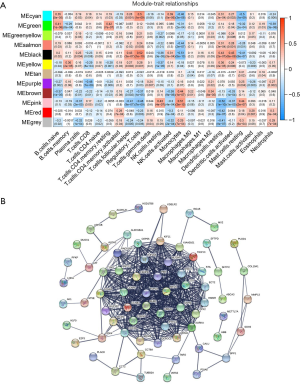
Next, we developed the PPI network (Figure 3B) using 111 module genes and eventually screened 36 candidate genes related to Tregs using the “CytoHubba” module of Cytoscape with all scores no less than 10.
Verification of the role of Treg-related genes in LUAD
To further verify the role of 36 candidate genes in LUAD, we extracted the gene sequencing and corresponding clinical data from the TCGA datvabase, comprising 375 LUAD and 48 normal samples. Using the Cox proportional hazards model, we eventually screened 13 hub genes (CCNB2, ECT2, RAD51AP1, UBE2C, CENPE, TOP2A, TYMS, KIF20A, STIL, CDKN3, PRC1, AURKA, HMMR) and their coefficients. Of the 13 hub genes, 4 (STIL, CCNB2, RAD51AP1, TOP2A) were considered risk genes (HRs: 0.2895–0.9717; all P<0.05), and their overexpression may lead to a worse prognosis. Additionally, 4 genes (HMMR, ECT2, TYMS, and PRC1) may serve as protective genes (HRs: 1.0030–2.6878; all P<0.05), and their overexpression may lead to a better outcome for LUAD. The results of Cox regression analyses are shown in detail in Table 1. The individual risk score of each LUAD patient was calculated according to our risk score formula. Based on the median risk score, the LUAD patients were assigned to the low- and high-risk groups.
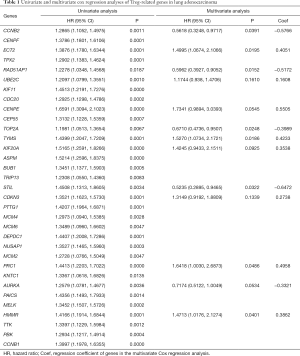
Full table
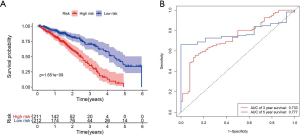
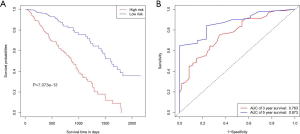
K-M survival curve analysis was performed to determine the difference between the two groups. The median survival time of the low-risk patients was 4.38 years, while that of the high-risk patients was 2.48 years (Figure 4A). The low-risk patients had a significantly better OS than the high-risk patients (3-year OS: 70.2% vs. 35.2%; 5-year OS: 36.6% vs. 0; P=1.651E-09), and the areas under the ROC curve (AUCs) were good (3-year AUC: 0.733; 5-year AUC: 0.777) (Figure 4B).
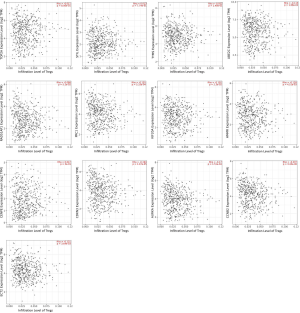
Spearman’s correlation between the Treg infiltration level and expression level of 13 hub genes were illustrated using the R package “ggstatsplot” (Figure 5). Of the 13 hub genes, we found that the expression levels of AURKA, CCNB2, CDKN3, ECT2, HMMR, KIF20A, PRC1, UBE2C, RAD51AP1, TOP2A and TYMS all had a positive correlation with Treg infiltration in the tumor microenvironment. By contrast, the correlation between the gene expression (of CENPE and STIL) and the infiltration level was negative.
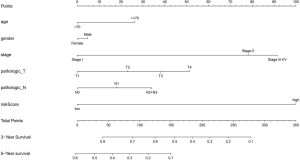
Prognostic nomogram model for LUAD: construction and validation
To apply Treg-related genes to clinical use, we constructed a nomogram model, combining the risk score with traditional clinical indicators, to predict the prognosis of LUAD. Given the high correlation between pathologic staging of M and T/N/stage, we finally included five clinical indicators (age, gender, stage, T and N) in the model (Figure 6).
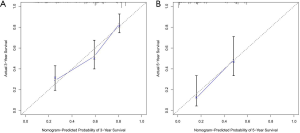
To test the discriminability of the model, we implemented the calibration curve, which is used to assess the accuracy of the (disease) risk model in predicting the probability of an individual outcome event in the future and reflects the degree of consistency between the predicted model risk and actual occurrence risk (23). In the study, the calibration curves showed that the predicted survival rate was consistent with the actual incidence rate within 3/5 years (Figure 7A,B). Moreover, we plotted the K–M curve, which showed good discriminating ability of the nomogram (P=7.073e−13) (Figure 8A), and the AUC was improved (3-year AUC: 0.763; 5-year AUC: 0.873) (Figure 8B).
GO and KEGG pathway enrichment analyses
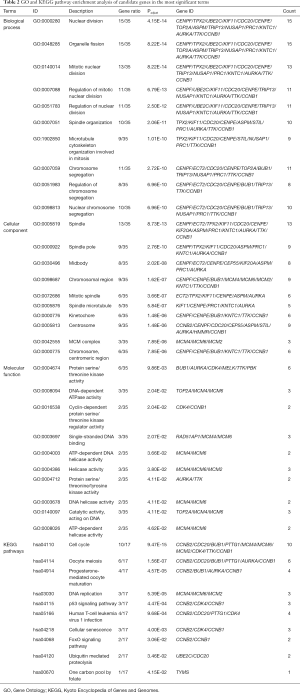
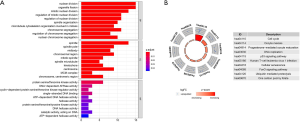
To identify molecular mechanisms of the candidate genes in LUAD, GO and KEGG pathway enrichment analyses were performed (Table 2). GO analysis includes 3 categories: biological processes (BP), cellular components (CC) and molecular function (MF). We found that the top enriched terms were (mitotic) nuclear division, spindle organization, organelle fission and chromosome segregation in BP; spindle, kinetochore and chromosome/centromeric region in CC; protein serine/threonine kinase activity, DNA-dependent ATPase activity and catalytic activity, acting on DNA in MF (Figure 9A). For KEGG enrichment pathways, the Treg-related hub genes were mostly enriched in the cell cycle, oocyte meiosis, DNA replication, p53 signaling pathway, human T-cell leukemia virus 1 infection, cellular senescence and ubiquitin-mediated proteolysis (Figure 9B).
Full table
Discussion
As the predominant histological phenotype of lung cancer, LUAD has a poor prognosis with a 5-year survival rate less than 25% (24,25), likely because of its undetected pathogenesis and complicated patterns of invasive growth, such as lymphovascular invasion, pleural invasion, and aerogenous invasion (26,27). Histopathological analysis revealed that the infiltration of inflammatory cells and lymphocytes is an important activity of the tumor microenvironment that may impact tumorigenesis, invasion, metastasis, and prognosis. Therefore, increasing emphasis has been placed on immune infiltration cell targeting compared with direct tumor cell killing (28). In recent years, immunotherapy for LUAD has advanced rapidly and has been markedly beneficial to patients (29). The 2018 Nobel Prize in physiology and medicine was awarded to Professor James P. Allison, and Professor Tasuku Honjo for their contribution to immune checkpoint therapy. To date, antibodies against PD-1/L1—nivolumab, pembrolizumab, cemiplimab, atezolizumab, durvalumab and avelumab—have been approved by the U.S. Food and Drug Administration (FDA) for first-line and/or late-stage treatment of 17 cancers (including NSCLC) (30,31). Pembrolizumab has been recommended for non-squamous cell carcinoma patients with high PD-L1 expression levels (tumor proportion score ≥50%) (32). However, the intratumor or inter-tumor heterogeneity and the non-standardized cut-off values for PD-1/L1, tumor mutation burden (TMB) and other independent immune-related biomarkers are far from being efficient (33-36). Thus, more potential and effective biomarkers are required.
In this study, we integrated three different GEO datasets (GSE10072/GSE32863/GSE43458) and obtained 196 LUAD and 137 normal samples. First, we acquired the abundances of 22 tumor-infiltrating immune cells using a deconvolution algorithm with the help of the CIBERSORTx portal, and then we identified candidate modules (black) and 36 candidate genes highly related to Tregs using the method of weighting gene coexpression network and PPIs. To verify the Treg-related genes, we generated the expression data of Treg-related genes as well as the clinical parameters from the TCGA database and then implemented a Cox proportional hazards model to calculate a risk score for each LUAD patient. The model performed well in that the low-risk patients had significantly longer 3- and 5-year survival times than the high-risk patients. Moreover, to further apply the Treg-related risk score in the clinic, we constructed a prognostic nomogram for LUAD, integrating the Treg-related risk score with traditional clinical parameters (age, gender, stage, T and N). The AUC value and calibration curve indicated that the nomogram performed better. Additionally, GO enrichment analysis revealed that the prognostic Treg-related genes were mainly enriched in BP involving cell proliferation, such as mitosis, chromosome separation, and DNA replicase activity. For the KEGG pathway analysis, Treg-related genes were mostly involved in the cell cycle, DNA replication and ubiquitin-mediated proteolysis, which were similar to the GO enriched terms for the cell cycle. This may reflect the reason for Treg enrichment in the tumor microenvironment, a finding that was consistent with that in previous studies (37-39). Moreover, the p53 and FoxO signaling pathways were included in the KEGG enriched pathways. P53 is a tumor suppressor that monitors the cell cycle, maintains genomic stability by participating in DNA repair and is coexpressed with angiogenic genes such as Smad4 to inhibit tumor angiogenesis (40,41). FoxO is a nuclear protein subfamily that mediates the inhibition of insulin or insulin-like growth factor to further influence cell cycle regulation, energy metabolism, protein stability, oxidative stress, apoptosis, and immunity (42,43).
Akimova et al. found that the number and inhibitory function of Treg intratumor were significantly higher than those in blood, lungs and lymph nodes by single-cell studies (37,38). Conventional research has highlighted the protective role of Tregs in alleviating inflammation in autoimmune diseases (44). Xie et al. showed that Tregs recruited to tumors played a role as “accomplices” in helping tumor cells escape immunological surveillance (45). Shimizu et al. and Marshall et al. revealed that the enrichment of Tregs in tumors usually indicates a poor prognosis (16,46). However, the latest discovery of Ferreira et al. found that Tregs promoted the differentiation of CD8+ effector memory T cells into tissue-resident memory T cells by providing the necessary cytokines, yielding more effective antitumor immunity (18). Therefore, studying the role of Treg cell infiltration into tumor tissue may provide a new perspective for immunotherapy or prognosis of LUAD.
Infiltrating Tregs in the tumor microenvironment play a potentially important role, which has been partially confirmed and applied in our study. However, this study has certain limitations. Although we drew the conclusion through multiple sequencing data and across different databases, more studies are needed to verify our results. Additionally, our results were obtained at the transcriptome level, and more proteomics level validation and clinical trials are needed to accelerate the clinical application.
Conclusions
We provide insights into the roles of Treg-related genes in LUAD and constructed a promising nomogram, which may help clinicians formulate more adequate treatment and follow-up plans.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 81772488).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-822
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-822). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Fitzmaurice C, Abate D, Abbasi N, et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol 2019;5:1749-68. [Crossref] [PubMed]
- Senosain MF, Massion PP. Intratumor Heterogeneity in Early Lung Adenocarcinoma. Front Oncol 2020;10:349. [Crossref] [PubMed]
- He R, Zuo S. A Robust 8-Gene Prognostic Signature for Early-Stage Non-small Cell Lung Cancer. Front Oncol 2019;9:693. [Crossref] [PubMed]
- Wang X, Yao S, Xiao Z, et al. Development and validation of a survival model for lung adenocarcinoma based on autophagy-associated genes. J Transl Med 2020;18:149. [Crossref] [PubMed]
- Peltanova B, Raudenska M, Masarik M. Effect of tumor microenvironment on pathogenesis of the head and neck squamous cell carcinoma: a systematic review. Mol Cancer 2019;18:63. [Crossref] [PubMed]
- Bremnes RM, Busund LT, Kilvaer TL, et al. The Role of Tumor-Infiltrating Lymphocytes in Development, Progression, and Prognosis of Non-Small Cell Lung Cancer. J Thorac Oncol 2016;11:789-800. [Crossref] [PubMed]
- Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene 2008;27:5904-12. [Crossref] [PubMed]
- Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science 2011;331:1565-70. [Crossref] [PubMed]
- Beyer M, Schultze JL. Regulatory T cells in cancer. Blood 2006;108:804-11. [Crossref] [PubMed]
- Ganesan AP, Johansson M, Ruffell B, et al. Tumor-infiltrating regulatory T cells inhibit endogenous cytotoxic T cell responses to lung adenocarcinoma. J Immunol 2013;191:2009-17. [Crossref] [PubMed]
- Domagala-Kulawik J, Osinska I, Hoser G. Mechanisms of immune response regulation in lung cancer. Transl Lung Cancer Res 2014;3:15-22. [PubMed]
- Vinay DS, Ryan EP, Pawelec G, et al. Immune evasion in cancer: Mechanistic basis and therapeutic strategies. Semin Cancer Biol 2015;35 Suppl:S185-98. [Crossref] [PubMed]
- Koyama K, Kagamu H, Miura S, et al. Reciprocal CD4+ T-cell balance of effector CD62Llow CD4+ and CD62LhighCD25+ CD4+ regulatory T cells in small cell lung cancer reflects disease stage. Clin Cancer Res 2008;14:6770-9. [Crossref] [PubMed]
- Wang W, Hodkinson P, McLaren F, et al. Small cell lung cancer tumour cells induce regulatory T lymphocytes, and patient survival correlates negatively with FOXP3+ cells in tumour infiltrate. Int J Cancer 2012;131:E928-37. [Crossref] [PubMed]
- Petersen RP, Campa MJ, Sperlazza J, et al. Tumor infiltrating Foxp3+ regulatory T-cells are associated with recurrence in pathologic stage I NSCLC patients. Cancer 2006;107:2866-72. [Crossref] [PubMed]
- Shimizu K, Nakata M, Hirami Y, et al. Tumor-infiltrating Foxp3+ regulatory T cells are correlated with cyclooxygenase-2 expression and are associated with recurrence in resected non-small cell lung cancer. J Thorac Oncol 2010;5:585-90. [Crossref] [PubMed]
- Erfani N, Mehrabadi SM, Ghayumi MA, et al. Increase of regulatory T cells in metastatic stage and CTLA-4 over expression in lymphocytes of patients with non-small cell lung cancer (NSCLC). Lung Cancer 2012;77:306-11. [Crossref] [PubMed]
- Ferreira C, Barros L, Baptista M, et al. Type 1 Treg cells promote the generation of CD8(+) tissue-resident memory T cells. Nat Immunol 2020;21:766-76. [Crossref] [PubMed]
- Rusk N. Expanded CIBERSORTx. Nat Methods 2019;16:577. [PubMed]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [Crossref] [PubMed]
- Jayaraman A, Jamil K, Khan HA. Identifying new targets in leukemogenesis using computational approaches. Saudi J Biol Sci 2015;22:610-22. [Crossref] [PubMed]
- Mattes MD, Weber WA, Foster A, et al. A Predictive Model for Lymph Node Involvement with Malignancy on PET/CT in Non-Small-Cell Lung Cancer. J Thorac Oncol 2015;10:1207-12. [Crossref] [PubMed]
- Huang YQ, Liang CH, He L, et al. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J Clin Oncol 2016;34:2157-64. [Crossref] [PubMed]
- Li Y, Tao L, Cai W. Profiles of Immune Infiltration and Prognostic Immunoscore in Lung Adenocarcinoma. Biomed Res Int 2020;2020:5858092. [PubMed]
- Zhang P, Li S, Lv C, et al. BPI-9016M, a c-Met inhibitor, suppresses tumor cell growth, migration and invasion of lung adenocarcinoma via miR203-DKK1. Theranostics 2018;8:5890-902. [Crossref] [PubMed]
- Shih AR, Mino-Kenudson M. Updates on spread through air spaces (STAS) in lung cancer. Histopathology 2020;77:173-80. [Crossref] [PubMed]
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Pilipow K, Darwich A, Losurdo A. T-cell-based breast cancer immunotherapy. Semin Cancer Biol 2020. Epub ahead of print. [Crossref] [PubMed]
- Reck M, Rodriguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. [Crossref] [PubMed]
- Topalian SL, Taube JM, Pardoll DM. Neoadjuvant checkpoint blockade for cancer immunotherapy. Science 2020;367:eaax0182. [Crossref] [PubMed]
- Santarpia M, Aguilar A, Chaib I, et al. Non-Small-Cell Lung Cancer Signaling Pathways, Metabolism, and PD-1/PD-L1 Antibodies. Cancers (Basel) 2020;12:1475. [Crossref] [PubMed]
- Hanna NH, Schneider BJ, Temin S, et al. Therapy for Stage IV Non-Small-Cell Lung Cancer Without Driver Alterations: ASCO and OH (CCO) Joint Guideline Update. J Clin Oncol 2020;38:1608-32. [Crossref] [PubMed]
- Greillier L, Tomasini P, Barlesi F. The clinical utility of tumor mutational burden in non-small cell lung cancer. Transl Lung Cancer Res 2018;7:639-46. [Crossref] [PubMed]
- Dempke WCM, Fenchel K, Dale SP. Programmed cell death ligand-1 (PD-L1) as a biomarker for non-small cell lung cancer (NSCLC) treatment-are we barking up the wrong tree? Transl Lung Cancer Res 2018;7:S275-9. [Crossref] [PubMed]
- Bai X, Wu DH, Ma SC, et al. Development and validation of a genomic mutation signature to predict response to PD-1 inhibitors in non-squamous NSCLC: a multicohort study. J Immunother Cancer 2020;8:e000381. [Crossref] [PubMed]
- Keenan TE, Burke KP, Van Allen EM. Genomic correlates of response to immune checkpoint blockade. Nat Med 2019;25:389-402. [Crossref] [PubMed]
- Akimova T, Zhang T, Negorev D, et al. Human lung tumor FOXP3+ Tregs upregulate four "Treg-locking" transcription factors. JCI Insight 2017;2:e94075. [Crossref] [PubMed]
- Ward-Hartstonge KA, Kemp RA. Regulatory T-cell heterogeneity and the cancer immune response. Clin Transl Immunology 2017;6:e154. [Crossref] [PubMed]
- Baratelli F, Lee JM, Hazra S, et al. PGE(2) contributes to TGF-beta induced T regulatory cell function in human non-small cell lung cancer. Am J Transl Res 2010;2:356-67. [PubMed]
- Tollini LA, Jin A, Park J, et al. Regulation of p53 by Mdm2 E3 ligase function is dispensable in embryogenesis and development, but essential in response to DNA damage. Cancer Cell 2014;26:235-47. [Crossref] [PubMed]
- Fan G, Tu Y, Chen C, et al. DNA methylation biomarkers for hepatocellular carcinoma. Cancer Cell Int 2018;18:140. [Crossref] [PubMed]
- Peixoto MS, da Silva FC Junior, de Oliveira Galvao MF, et al. Oxidative stress, mutagenic effects, and cell death induced by retene. Chemosphere 2019;231:518-27. [Crossref] [PubMed]
- Wu Y, Peng H, Cui M, et al. CXCL12 increases human neural progenitor cell proliferation through Akt-1/FOXO3a signaling pathway. J Neurochem 2009;109:1157-67. [Crossref] [PubMed]
- Miyara M, Ito Y, Sakaguchi S. TREG-cell therapies for autoimmune rheumatic diseases. Nat Rev Rheumatol 2014;10:543-51. [Crossref] [PubMed]
- Xie M, Wei J, Xu J. Inducers, Attractors and Modulators of CD4(+) Treg Cells in Non-Small-Cell Lung Cancer. Front Immunol 2020;11:676. [Crossref] [PubMed]
- Marshall EA, Ng KW, Kung SH, et al. Emerging roles of T helper 17 and regulatory T cells in lung cancer progression and metastasis. Mol Cancer 2016;15:67. [Crossref] [PubMed]



