
Circ-AASDH functions as the progression of early stage lung adenocarcinoma by targeting miR-140-3p to activate E2F7 expression
Introduction
Non-small cell lung cancer (NSCLC) accounts for approximately 85% of lung cancers, and lung adenocarcinoma (LUAD) is the most common subtype (1,2). Multi-modal therapy, including surgery, radiotherapy, chemotherapy and targeted therapy, is the primary treatment for LUAD patients (3). However, most LUAD patients cannot be timely detected in the early stage because of lacking the effective diagnostic tools and even developed to the middle and advanced clinical stage. Although the prognosis of LUAD is better than other subtypes, the 5-year survival rate is still less than 20% (4). Therefore, it is urgently needed to find more specific and sensitive indicators to monitor the prognosis and recurrence of LUAD.
CircRNAs, as novel non-coding RNAs, which were formed by back-splicing exons and/or introns without 5' and 3', can be divided into exonic circRNAs (EciRNA), circular intronic RNAs (ciRNAs) and exon-intron circRNA (EIciRNA) and had been proved to exist widely in various tissues (5-8). Recently, circRNAs have attracted more and more attention because of its involvement in the occurrence and progression of malignant tumors at transcriptional and translation levels (9). As the focus of ceRNA research, recent studies have shown that circRNAs are rich in microRNA (miRNA) binding sites and play the role of miRNA sponge in cells, thus relieving the target genes inhibition (10). Hsa_circ_001350, for example, regulated glioma cell proliferation, apoptosis, and metastatic properties by sponging miR-1236 (11). Circ-LDLRAD3 served as an oncogene to promote NSCLC progression through upregulating SLC1A5 via sponging miR-137 (12). Circ-AASDH (hsa_circ_0126678), located at chr4: 57237616-57250507 with a length of 903 bp, is shaped via back-splicing of 2–4 exons of pre-AASDH. According to the GSE101684, a GEO dataset contained microarray data of circRNAs expression of LUAD, we found circ-AASDH was strongest aberrantly expressed circRNA among all candidate genes. Therefore, circ-AASDH was selected as the target molecule to analysis mechanism and further function in LUAD.
In our research, we proved that circ-AASDH was up-regulated in LUAD cells and tissues and highly expressed circ-AASDH correlated with tumor size, clinical stage and poor prognosis. In function, we found that circ-AASDH promoted tumor growth in vivo, and knockdown of circ-AASDH inhibit the proliferation, invasion of LUAD in vitro. Next, we observed that circ-AASDH promoted the expression of E2F Transcription Factor 7 (E2F7) via sponging miR-140-3p. Our results discovered a new regulatory mechanism of circ-AASDH in LUAD and provided a promising therapeutic target for patients with LUAD.
We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-1062).
Methods
Patient tissues
The specimens of 59 patients with LUAD were collected without any radiotherapy or chemotherapy before surgical operation. Immediately freeze all samples in liquid nitrogen and store them at −80 °C until RNA is extracted. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the Ethics Committee of the Affiliated Hospital of Qingdao University (QYFYKY 2018-10-11-2), and all patients signed informed consent forms.
Cell culture and transfection
All LUAD cell lines, including A549, H1299, H524, H332, H1650 and bronchial epithelium transformed with Ad12-SV40 2B (BEAS-2B) were cultured in RPMI1640 with 10% FBS. All the above cells grew in the incubator at 37 °C containing 5% CO2.
SiRNAs were used to knock down circ-AASDH expression and si-normal control (si-NC) was used as a control, which were purchased from Sangon Biotech (Shanghai, China). The siRNAs sequences were manifested in Table 1. Anti-miR-140-3p (miR-140-3p inhibitor), and its normal control (anti-miR-NC), over-expressed miR-140-3p (miR-140-3p) and its matched normal control (miR-NC), were synthesized by Sangon Biotech (Shanghai, China). In order to construct the over-expressed circ-AASDH plasmid (circ-AASDH), the circ-AASDH full-length complementary DNA (cDNA) was inserted into the pLCDH-CIR vector (BGI, Qingdao, China), the normal vector was used as a control. According to the manufacturer's instructions, use Polyplus-transfection® (Illkirch, France) to transfect LUAD cells with these plasmids or RNAs at appropriate doses.

Full table
Verification of circular structure
In order to verify the circular structure of circ-AASDH, the linear transcripts and circular transcripts of AASDH were strengthened from genomic DNA (gDNA) and cDNA of LUAD cells by convergent primers and primers, respectively. Ideally, circ-AASDH in cDNA can be amplified by divergent primers through isolating PCR products by agarose gel, but not by gDNA. In addition, the loop junction was sequenced to confirm the circ-AASDH sequence. Meanwhile, RNase R was used to perform the linear and circular AASDH expression levels in LUAD cells.
RNA extraction and qRT-PCR
TRIzol reagent (Haigene, Haerbin, China) was applied to extract total RNA from tissue and cell. CDNA was obtained by cDNA synthesis kit (Takara, Beijing, China). The SYBR Green PCR Kit (Takara, Beijing, China) was performed for qPCR with circRNA, miRNA, mRNA and the primers (Tsingke, Qingdao, China) were listed in Table 1. We used 2-ΔΔCt method to calculate and analyze the relative expression, GAPDH as the internal control of circRNA and mRNA and U6 was used to miRNA.
Cell proliferation and colony formation assay
LUAD cells viability was evaluated through CCK-8 Kit (Dojindo, Tokyo, Japan). The transfected cells (1×104/well) were inoculated into 96-well plates. After incubation for 1 day, 2 days, 3 days under suitable condition, 10 µL CCK8 solution was added for 4h at 37 °C. The microplate spectrophotometer (Winooski, VT, USA) was used to measure the optical density of each hole under 450 nm. As described in the previous article (13), 1×103 transfected cells were cultured for 14 days with 6-well plate, and then the colonies were counted after treating methanol and Giemsa.
Wound-healing assay
After transfection, the cells were cultured into a 6-well plate until they were reconnected and 80% sub-confluence was reached. The single-cell layer was scratched with the tip of a 10 µL pipette and then washed 3 times with 1×PBS to clear the cell fragments and add fresh FBS. The images were acquired at 0 and 48 h, and the relative width of the wound was calculated by ImageJ software.
Migration assays
Transwell assay was applied to evaluate the migrant ability of LUAD cells in 8 µL aperture and polycarbonate filter Boyden chamber (Corning, New York, USA). In brief, the medium with 10% FBS was used in the lower chamber, while the upper chamber with the free medium. After culturing at 5% CO2 with 37 °C for 24 h, 70% ethanol and stained with 0.1% crystal violet need fixing with the cells, above cells were measured by microscope (Nikon Ti-s, Shanghai, China).
Flow cytometry analysis
Apoptosis rates of transfected A549 and H1299, according to the manufacturer’s instructions, were performed by the Annexin-V FITC Apoptosis Kit (Invitrogen, Beijing, China). Cell apoptosis was detected by ACCURI C6 flow cytometry (BD, CA, USA).
Western blot assay
The total proteins of LUAD cells were isolated by ExKine Cytoplasmic Protein Extraction Kit (Abbkine, CA, USA) and quantified with BCA protein Assay Kit (Thermo scientific). The membrane containing 5% skimmed milk was used to seal 60 min at room temperature and incubated with the following antibodies: β-actin (Univ, Shanghai, China) and anti-E2F7 (Univ, Shanghai, China). Then, the membrane was incubated with second antibodies and photographed.
Luciferase reporter assay
In order to identify the binding sites between circ-AASDH and miR-140-3p, the circ-AASDH wild type or mutant sequence containing miR-140-3p binding sites was inserted into psiCHECK2 vector, labeled as circ-AASDH-wt and circ-AASDH-mut, respectively. Later, miR-140-3p or miR-NC was co-transfected with the luciferase vector into LUAD cells. After transfection 48 h, luciferase activity was performed by Luciferase Reporter Assays System (Promega, Beijing, China). Above procedures were also carried out to verify the binding between E2F7 and miR-140-3p.
RNA pull-down assay
Non-specific negative control (Bio-miR-NC), mutant and wild-type biotin-labeled miR-140-3p were labeled as Bio-miR-140-3p-mut and Bio-miR-140-3p-wt, were transfected into LUAD cells. After 2 days, the cell lysate was combined with streptavidin-labeled magnetic beads, and then the circ-AASDH expression level was performed to measure in RNA-complex.
Xenografts in mice
Experiments were performed under a project license (NO. 19-256A) granted by Laboratory Animal Center of Qingdao University, in compliance with institutional guidelines for the care and use of animals. All nude mice obtained from Laboratory Animal Center of Qingdao University (5 mice per group age, weight 18 to 22 g, about 4 weeks). About 2×105 stably encoding circ-AASDH A549 cells and normal control vector were subcutaneously injected into the lower part of the armpit. The volume and weight of the tumor were assessed per 5 days, and the body weight was checked until the mice were dissected.
Bioinformatics analysis
We used the GEO database (https://www.ncbi.nlm.nih.gov/geo/) to screen differentially expressed circRNAs, and the targets miRNAs of selected circRNA were predicted by online website, circBank (http://www.circbank.cn/searchMiRNA.html) and CircInteractome (https://circinteractome.nia.nih.gov/mirna_target_sites.html). RNA22 (https://cm.jefferson.edu/rna22/), TargetScan (http://www.targetscan.org/vert_72/), PicTar (https://pictar.mdc-berlin.de/), miRmap (https://mirmap.ezlab.org/) and mircoT-CDS (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index) were used to speculate the targets of miR-140-3p.
Statistical analysis
All data were performed and evaluated through SPSS software (version 23.0), R software (version 4.0.2) and GraphPad Prism software (version 8.0), the statistically significant index was designated as P<0.05. Differences in two groups were assessed as average ± SD, and Chi-square test was measured to assess the differences of categorical variables.
Results
Identification circ-AASDH expression in LUAD tissues and cells
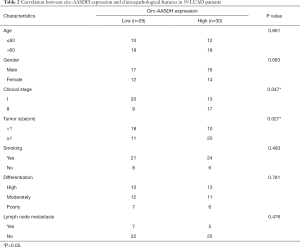
We analyzed the GSE101684 microarray, containing four LUAD tissues and four normal tissues, to obtain the differentially expressed circRNA (DEcirc) in tissues. The screening criteria were as follows: the absolute value of logFC >1, P<0.05 as the significant differences between the two groups (Figure 1A). Heatmap showed the top ten up-regulated and down-regulated circular RNAs in GSE101486 (Figure 1B). Hsa_circ_0126678, also named hsa_circ_406483, was discovered the most DEcircRNA and had not been studied in LUAD yet. It is 903 bp in length and comes from 2–4 exons of Aminoadipate-Semialdehyde Dehydrogenase (AASDH) located on chromosome 4, thus we called it as circ-AASDH. Subsequently, the back-spliced region sequence of circ-AASDH was identified by Sanger sequencing (Figure 1C). As shown in Figure 1D, the expression of circ-AASDH was significantly higher in human LUAD cells (A549, H1299, H524, H322 and H1650) than that in human normal pulmonary epithelial cell line BEAS-2B. In addition, as shown in Figure 1E, liner mRNA AASDH, can be produced by convergent primers from cDNA and gDNA of A549 cell, but circRNA AASDH can not be amplified by gDNA. Meanwhile, GAPDH can be amplified in gDNA and cDNA, while circ-GAPDH cannot. After treatment with RNase R, the expression of mRNA AASDH was significantly lower than that of circRNA AASDH (Figure 1F,G). Subsequently, the expression level of circ-AASDH in LUAD tissues and normal adjacent tissue samples from 59 LUAD patients was elevated (Figure 1H). Noticeably, the increased expression of circ-AASDH was found to be involved in tumor size and clinical stage (Table 2). Furthermore, we evaluated the relationship between overall survival (OS) and expression of circ-AASDH. As exhibited in Figure 1I, the OS of LUAD patients with the high group of circ-AASDH expression were poor in comparison with that in the low group. The above results demonstrated the circ-AASDH expression was up-regulated in LUAD and the circular structure of circ-AASDH.
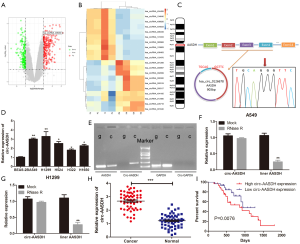
Full table
Silencing of circ-AASDH inhibited LUAD progression in vitro
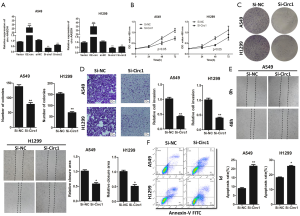
To reveal the outcomes of circ-AASDH in LUAD progression, two siRNAs targeting of circ-AASDH (si-circ-AASDH #1 and si-circ-AASDH #2) were synthesized and transfected into LUAD cells. At the same time, the circ-AASDH-overexpressing vector (OE-circ-AASDH) was constructed into LUAD cells. As shown in Figure 2A, circ-AASDH expression was significantly increased and inhibited in A549 and H1299 cells after transfecting plasmid and siRNA. Owe to si-circ-AASDH #1 has the highest efficiency to knockdown, and it was applied in subsequent experiments. The silencing of circ-AASDH could inhibit the cell proliferation ability of LUAD cells (Figure 2B). Equivalently, there were fewer clones verified the anti-proliferative ability of lower circ-AASDH groups in LUAD cells (Figure 2C). As demonstrated by migration assays, down-regulated circ-AASDH could lead to a decline of migrating ability in LUAD cells (Figure 2D). Consistently, the cells with circ-AASDH inhibition also revealed defects in wound healing compared to the normal control (Figure 2E). circ-AASDH depletion promoted apoptosis in A549 and H1299 cells by flow cytometry analysis with Annexin-V/PI (Figure 2F). Taken together, these findings demonstrated that silencing of circ-AASDH could suppress LUAD cell proliferation and migration, and induce apoptosis.
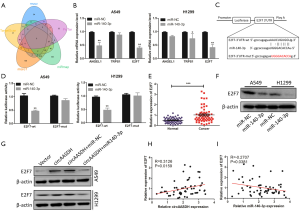
Circ-AASDH acted as sponging for miR-140-3p in LUAD cells
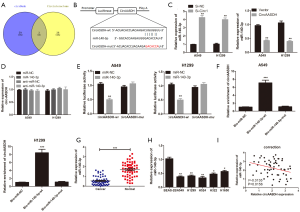
The CircInteractome and circBank databases were used to search the miRNAs of circ-AASDH for exploring the molecular mechanism in LUAD progression. The Venn diagram displayed hsa-miR-548k, hsa-miR-891b and hsa-miR-140-3p by overlapping the prediction results (Figure 3A). MiR-140-3p was detected down-regulated in LUAD, according to the online database from dbDEMC (https://www.picb.ac.cn/dbDEMC/index.html). Therefore, miR-140-3p was chosen as the putative downstream of circ-AASDH. Figure 3B suggested that the binding sites between miR-140-3p and circ-AASDH, and the mutant sequence of circ-AASDH. What’s more, the knockdown of circ-AASDH could promote miR-140-3p expression, while OE-circ-AASDH reduced the expression of miR-140-3p in A549 and H1299 cells (Figure 3C). However, the level of circ-AASDH would not change because of miR-140-3p knockdown or up-regulation (Figure 3D). Subsequent luciferase reporter assays clarified that compared with the miR-NC group, miR-140-3p transfection can reduce the luciferase activity of the circ-AASDH-wt. Nevertheless, between the miR-NC group and the miR-140-3p group, there was no significant change in luciferase activity of circ-AASDH-mut group (Figure 3E). RNA pull-down assay revealed that Bio-miR-140-3p-wt captured more circ-AASDH than Bio-miR-NC in LUAD cells, but it doesn’t influence when the binding site on circ-AASDH was mutant (Figure 3F). As expected, the expression of miR-140-3p in LUAD tissue specimens and cell lines was confirmed to be decreased (Figure 3G,H). Besides, the scatter plot showed that the level of miR-140-3p in LUAD was negatively correlated with the expression of circ-AASDH (Figure 3I). In conclusion, circ-AASDH inhibited the expression of miR-140-3p in LUAD tissues and cells by serving as a molecular sponge.
Circ-AASDH antagonizes the inhibitory effect of miR-140-3p on the progression of LUAD in vitro
To further understand the functions of miR-140-3p and research whether miR-140-3p was related to CircASSDH in LUAD, we co-transfected LUAD cells with miR-140-3p and circ-AASDH, or transfected miR-140-3p into A549 and H1299 cells alone. As depicted in Figure 4A, after transfecting with miR-140-3p mimics, the miR-140-3p expression level was significantly up-regulated in A549 and H1299 cells, while the addition of circ-AASDH reversed the effect. In function, the miR-140-3p mimics could significantly inhibit cell viability (Figure 4B) and colony formation ability (Figure 4C), while the up-regulation of circ-AASDH weakened this anti-proliferation effect. Similarly, miR-140-3p mimics significantly repressed cell migrant ability, but this effect was neutralized after co-transfection with circ-AASDH (Figure 4D). Additionally, overexpression of miR-140-3p resulted in lower cell proliferation effect (Figure 4E). In general, circ-AASDH facilitated the malignant phenotype of LUAD via miR-140-3p sponge.
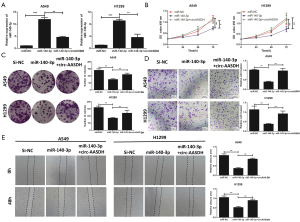
E2F7 expression was facilitated by miR-140-3p in LUAD cells
To explore the biological mechanism of miR-140-3p in the progress of LUAD, we used five online databases to predict the target genes of miR-140-3p, which are RNA22, PicTar, Mirmap, MircoT, and Targetscan. According to Venn diagram, three putative targets (ANGEL1, TRPS1 and E2F7) were screened (Figure 5A). Subsequent qRT-PCR showed that effects of miR-140-3p on the level expressions of above genes were explored in A549 and H1299 cells. The Figure 5B exhibited that only E2F7 expression was both decreased in LUAD cells transfected with miR-140-3p. Hence, E2F7 was chosen as the target gene for next researches. The binding site between miR-140-3p and the 3'UTR of E2F7 was predicted, and their relationship were proved by the luciferase reporter assays (Figure 5C,D). The qRT-PCR data further confirmed the up-regulated E2F7 in LUAD tissues compared with adjacent normal tissues (Figure 5E). MiR-140-3p mimics could inhibit the E2F7 protein level in A549 and H1299 cells through Western blotting analysis (Figure 5F). As exhibited in Figure 5G, the E2F7 protein expression increased in transfected OE-circ-AASDH A549 and H1299 cells, and this effect was weakened through the recovery of miR-140-3p expression. Scatter plot depicted that the expression of E2F7 in LUAD was positively correlated with circ-AASDH and negatively correlated with miR-140-3p (Figure 5H,I). These results support that circ-AASDH could up-regulate the expression level of E2F7 in LUAD cells by sponging miR-140-3p.
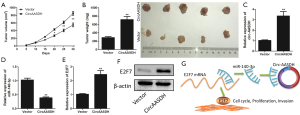
Circ-AASDH promoted tumor growth in vivo
In order to further verify the circ-AASDH function in the development of LUAD in vivo, A549 cells with stably OE-circ-AASDH were injected into nude mice to establish the xenograft tumor models. As shown in Figure 6A, the knockdown of circ-AASDH significantly reduced the growth of tumor. Similarly, the weight of tumors derived from the OE-circ-AASDH group was significantly increased compared to the vector (Figure 6B). As expected, compared with the vector group, the mRNA levels of circ-AASDH and E2F7 in the resected tumor tissue in the OE-circ-AASDH group increased, while the expression of miR-140-3p decreased (Figure 6C,D,E). In addition, OE-circ-AASDH caused an increase in E2F7 protein levels (Figure 6F). These findings indicated that the high expression of circ-AASDH promoted the growth of LUAD in vivo.
Discussion
It is noteworthy that the development of modern medical imaging technology and the increasing popularity of physical examination can result in a distinct improvement of the sensitivity and specificity in the diagnosis of early-stage lung cancer, but there are still some false negative which need to be screened by early biomarkers (14). Circular RNA has draw wide attention in recent years because of its high stability, especially in the early diagnosis of human cancer (5). Nevertheless, the exact roles of circRNA in LUAD are still undiscovered. Deeply revealing the mechanism of circRNA is essential to discover therapeutic targets and promising biomarkers in LUAD patients. We chose circ-AASDH from the GSE101486 dataset in the GEO database to clarify its biological significance and potential mechanism in LUAD. Our findings suggested that circ-AASDH acted as a sponge for miR-140-3p to reduce its inhibitory effect on E2F7, thereby promoting progression in LUAD.
So far, more and more circRNAs have been found to be related to pathophysiological events in LUAD, such as hsa_circRNA_002178 could enhance PD-L1 expression by sponging miR-34 in LUAD to induce T-cell exhaustion (15). Circ_EPB41L2 acted as a tumor inhibitor to suppress the migration invasion and proliferation, through regulating CDH4 expression by miR-211-5p in LUAD cells (16). Hsa_circ_0006427 suppressed the progression of LUAD through regulating miR-6783-3p-DKK1 axis and inhibiting Wnt/β-catenin pathway (17). In this research, circ-AASDH was identified as circRNA by RNase R processing, PCR, and Sanger sequencing. The expression of circ-AASDH was enhanced in LUAD tissue and cell. In addition, the high expression of circ-AASDH was involved in tumor size, clinical stage and poor prognosis. Silencing of circ-AASDH can inhibit the proliferation and migration of LUAD cells and enhance apoptosis. These findings indicated that circ-AASDH is carcinogenic in LUAD. Although circ-AASDH has not been studied in other cancers, researchers will continue to explore its function.
Recently, circRNA, known as competitive endogenous RNA (ceRNA), can affect miRNA expression, thus reducing its inhibition on target genes (18). Most exons circRNA exist in the cytoplasm, and circ-AASDH is no exception, suggesting that it may play a role by regulating post-transcription. Therefore, we predicted that circ-AASDH may participate in the regulation of LUAD via similar ceRNA mechanisms. According to the prediction of bioinformatics websites and the results of luciferase reporter and RNA pull-down assay, miR-140-3p was examined to be the direct target of circ-AASDH. It has been previously proved that miR-140-3p has a tumor suppressive effect in some types of human malignant tumors. Ovarian cancer (19), for instance, spinal chordoma (20), cutaneous melanoma (21), breast cancer (22) and NSCLC (23). In this research, the expression of miR-140-3p was down-regulated in LUAD tissues and cells, and negatively correlated with the expression of circ-AASDH. In mechanism, miR-140-3p mimics could cell proliferation and migration, suggesting that miR-140-3p had an anti-tumor effect in LUAD. However, the inhibition of cell proliferation and migration induced by miR-140-3p was significantly counteracted by the overexpression of circ-AASDH. Based on the above results, we demonstrated that circ-AASDH promoted the LUAD process by sponging miR-140-3p.
E2F Transcription Factor 7 (E2F7) was studied to play essential roles in the regulation of cell cycle progression (24-26). Also, E2F7 is involved in the regulation of carcinogenic signal pathway and the progression of tumors (27,28). For example, E2F7 expression of prostate cancer tissues was found to be notably higher compared with adjacent normal tissues (27). In the present study, according to the results of luciferase reporter and western blot assay, we found that E2F7 was the target gene of miR-140-3p in LUAD cells, and E2F7 was highly expressed in LUAD tissues. Furthermore, the overexpression of circ-AASDH led to increased E2F7 expression, while the recovery of miR-140-3p weakened this effect. Moreover, the expression of E2F7 was positively correlated with circ-AASDH, but negatively related to miR-140-3p in LUAD tissues. Therefore, it is concluded that circ-AASDH acted as a miR-140-3p sponge to eliminate its inhibitory effect on E2F7. In short, circ-AASDH promoted the LUAD procession via sponging miR-140-3p and activating E2F7. Our study illustrated the circ-AASDH/miR-140-3p/E2F7 regulating axis in LUAD (Figure 6G). Moreover, circRNAs still have many putative functions to be explored, such as regulating gene transcription and acting as a translation template for proteins, regulating the stability of mRNA, and interacting with RNA-binding proteins (6,29). Other possible molecular mechanisms of circ-AASDH in LUAD will be further studied in subsequent research.
Conclusions
To sum up, circ-AASDH was up-regulated in LUAD cells and tissues. Highly expressed circ-AASDH was involved in tumor size, clinical stage and poor prognosis. Further experiments revealed that circ-AASDH regulated the expression of E2F7 through sponging miR-140-3p, which contributes to the malignant progression of LUAD. Our results provide ideas for understanding the signaling pathways of circ-AASDH/miR-140-3p/E2F7 axis and clarify potential therapeutic targets in LUAD. However, due to the diversification of circRNA functions, most of the functions of circ-AASDH still need to be explored, more functional demonstration and related signal pathways need to be further studied.
Acknowledgments
We thank AME Editing Service, (http://editing.amegroups.cn/#editing) for its linguistic assistance during the preparation of this manuscript.
Funding: This study was funded by Key R & D programs in Shandong Province (grant number: 2018GSF118119).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1062
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-1062
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-1062). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the Ethics Committee of the Affiliated Hospital of Qingdao University (QYFYKY 2018-10-11-2), and all patients signed informed consent forms. Animal experiments were performed under a project license (NO.: 19-256A) granted by Laboratory Animal Center of Qingdao University, in compliance with its institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Takamori S, Shimokawa M, Matsubara T, et al. Prognostic Impact of Smoking Period in Patients with Surgically Resected Non-small Cell Lung Cancer. Ann Surg Oncol 2021;28:685-94. [Crossref] [PubMed]
- Ben Dori S, Aizic A, Sabo E, et al. Spatial heterogeneity of PD-L1 expression and the risk for misclassification of PD-L1 immunohistochemistry in non-small cell lung cancer. Lung Cancer 2020;147:91-8. [Crossref] [PubMed]
- Jiang K, Zhi X, Shen Y, et al. Impact of examined lymph node count on long-term survival of T1-2N0M0 double primary NSCLC patients after surgery: a SEER study. PeerJ 2020;8:e8692. [Crossref] [PubMed]
- Yin Y, Long J, He Q, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer 2019;10:5015-21. [Crossref] [PubMed]
- Wang Y, Lu T, Wang Q, et al. Circular RNAs: Crucial regulators in the human body Oncol Rep 2018;40:3119-35. (Review). [Crossref] [PubMed]
- Zhou R, Wu Y, Wang W, et al. Circular RNAs (circRNAs) in cancer. Cancer Lett 2018;425:134-42. [Crossref] [PubMed]
- Wu P, Mo Y, Peng M, et al. Emerging role of tumor-related functional peptides encoded by lncRNA and circRNA. Mol Cancer 2020;19:22. [Crossref] [PubMed]
- Liu J, Zhang X, Yan M, et al. Emerging Role of Circular RNAs in Cancer. Front Oncol 2020;10:663. [Crossref] [PubMed]
- Ren S, Lin P, Wang J, et al. Circular RNAs: Promising Molecular Biomarkers of Human Aging-Related Diseases via Functioning as an miRNA Sponge. Mol Ther Methods Clin Dev 2020;18:215-29. [Crossref] [PubMed]
- Liu Y, Ma C, Qin X, et al. Circular RNA circ_001350 regulates glioma cell proliferation, apoptosis, and metastatic properties by acting as a miRNA sponge. J Cell Biochem 2019;120:15280-7. [Crossref] [PubMed]
- Xue M, Hong W, Jiang J, et al. Circular RNA circ-LDLRAD3 serves as an oncogene to promote non-small cell lung cancer progression by upregulating SLC1A5 through sponging miR-137. RNA Biol 2020;17:1811-22. [Crossref] [PubMed]
- Wang Y, Xu R, Zhang D, et al. Circ-ZKSCAN1 regulates FAM83A expression and inactivates MAPK signaling by targeting miR-330-5p to promote non-small cell lung cancer progression. Transl Lung Cancer Res 2019;8:862-75. [Crossref] [PubMed]
- Millares L, Barreiro E, Cortes R, et al. Tumor-associated metabolic and inflammatory responses in early stage non-small cell lung cancer: Local patterns and prognostic significance. Lung Cancer 2018;122:124-30. [Crossref] [PubMed]
- Wang J, Zhao X, Wang Y, et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis 2020;11:32. [Crossref] [PubMed]
- Zhang SJ, Ma J, Wu JC, et al. CircRNA EPB41L2 inhibits tumorigenicity of lung adenocarcinoma through regulating CDH4 by miR-211-5p. Eur Rev Med Pharmacol Sci 2020;24:3749-60. [PubMed]
- Yao Y, Hua Q, Zhou Y. CircRNA has_circ_0006427 suppresses the progression of lung adenocarcinoma by regulating miR-6783-3p/DKK1 axis and inactivating Wnt/beta-catenin signaling pathway. Biochem Biophys Res Commun 2019;508:37-45. [Crossref] [PubMed]
- Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer 2018;17:79. [Crossref] [PubMed]
- Wang J, Zhu M, Zhou X, et al. MiR-140-3p inhibits natural killer cytotoxicity to human ovarian cancer via targeting MAPK1. J Biosci 2020;45:66. [Crossref] [PubMed]
- Zou MX, Huang W, Wang XB, et al. Identification of miR-140-3p as a marker associated with poor prognosis in spinal chordoma. Int J Clin Exp Pathol 2014;7:4877-85. [PubMed]
- He Y, Yang Y, Liao Y, et al. miR-140-3p Inhibits Cutaneous Melanoma Progression by Disrupting AKT/p70S6K and JNK Pathways through ABHD2. Mol Ther Oncolytics 2020;17:83-93. [Crossref] [PubMed]
- Zhou Y, Wang B, Wang Y, et al. miR-140-3p inhibits breast cancer proliferation and migration by directly regulating the expression of tripartite motif 28. Oncol Lett 2019;17:3835-41. [Crossref] [PubMed]
- Hu C, Zou Y, Jing LL. miR-140-3p inhibits progression of non-small cell lung cancer by targeting Janus kinase 1. J Biosci 2020;45:48. [Crossref] [PubMed]
- Di Stefano L, Jensen MR, Helin K. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J 2003;22:6289-98. [Crossref] [PubMed]
- Mitxelena J, Apraiz A, Vallejo-Rodriguez J, et al. An E2F7-dependent transcriptional program modulates DNA damage repair and genomic stability. Nucleic Acids Res 2018;46:4546-59. [Crossref] [PubMed]
- Westendorp B, Mokry M, Groot Koerkamp MJ, et al. E2F7 represses a network of oscillating cell cycle genes to control S-phase progression. Nucleic Acids Res 2012;40:3511-23. [Crossref] [PubMed]
- Wang Y, Pei X, Xu P, et al. E2F7, regulated by miR30c, inhibits apoptosis and promotes cell cycle of prostate cancer cells. Oncol Rep 2020;44:849-62. [Crossref] [PubMed]
- Yin W, Wang B, Ding M, et al. Elevated E2F7 expression predicts poor prognosis in human patients with gliomas. J Clin Neurosci 2016;33:187-93. [Crossref] [PubMed]
- Shi Y, Jia X, Xu J. The new function of circRNA: translation. Clin Transl Oncol 2020;22:2162-9. [Crossref] [PubMed]


