
A machine learning-based prediction of the micropapillary/solid growth pattern in invasive lung adenocarcinoma with radiomics
Introduction
Lung adenocarcinoma is established to be the most common subtype of lung cancer, with high heterogeneity in its molecular, pathological, and clinical aspects. Based on the histological pattern, a classification by the International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society further categorized invasive lung adenocarcinoma into the five following subtypes: lepidic, acinar, solid, papillary, and micropapillary (1). It is now required that each histological pattern and its percentage be recorded in the pathological reports. Many studies (2-4) have supported the histological classification of lung adenocarcinoma being correlated with patient’s prognosis, lymph node metastasis, and epidermal growth factor receptor (EGFR) mutation status. Patients with the micropapillary (MP)-predominant and solid (S)-predominant subtype are considered to have worse survival prognosis and higher recurrence rates. Studies also have shown that even a MP component of <5% has a significant prognostic impact on patient’s survival (5,6). Moreover, patient with an MP component >5% undergoing limited resection, but not lobectomy, would carry a higher risk of tumor recurrence compared with those having an MP component of <5% (7). Therefore, preoperative predictions of the presence of an MP/S component can provide important information for deciding a patient’s surgical strategies.
Radiomics (often called texture/shape analysis) can be used to characterize the medical images via quantitative image analysis; thus, it may represent a valuable source of information for lesion diagnosis and prognosis prediction (8). Machine learning, an artificial intelligence subfield, can utilize radiomics features to automatically infer decisions from the datasets with the purpose of developing a decision-making model. In regrading lung cancer, computed tomography (CT)-based radiomics are a practical, low-cost tool for noninvasively characterizing lung lesions and discerning between malignant and benign lung nodules (9), thus stratifying the risk of mediastinal lymph node metastasis (10) and discriminating invasive from indolent adenocarcinomas (11). However, highly accurate and reliable machine learning methods are still lacked in driving the success of radiomics applications in clinical care, and should be chosen appropriately (12).
Thus, in our study, we aimed to evaluate and compare the predictive value of radiomics-based machine learning models in the presence of an MP/S growth pattern of lung adenocarcinoma.
We present the following article in accordance with the TRIPOD reporting checklist (available at http://dx.doi.org/10.21037/tlcr-21-44).
Methods
This was a multicenter study with patients retrospectively included from three different institutions, including Shanghai Pulmonary Hospital, Ningbo No. 2 Hospital, and the Affiliated Hospital of Zunyi Medical College. The waiver of informed consent was approved by the institutional review board (IRB) of each participating hospital. The study was performed in accordance with the Declaration of Helsinki (as revised in 2013). The complete design of our study is illustrated in Figure 1.

Study population
Patients undergoing resection for invasive lung adenocarcinoma from January 2011 to December 2013 at Shanghai Pulmonary Hospital, and from January 2013 to December 2014 in the other two hospitals were retrospectively recruited in this study. The inclusion criteria were as follows: (I) pathologically confirmed invasive lung adenocarcinoma; (II) complete preoperative thin-section CT image (1 mm) data with follow-up survival data; and (III) tumor size measured in CT images <3 cm. We excluded those patients with multifocal lesions, neoadjuvant chemotherapy or radiotherapy. The dataset from Shanghai Pulmonary Hospital with the most included patients (N=268) was used as the primary cohort for model development, and the datasets (N=193) from the other two hospitals were used as independent external validation cohorts.
All patients were followed up every 6 months for the first 3 years after surgery and every year thereafter. Follow-up examinations included thoracic CT scans, carcinoembryonic antigen (CEA) levels, abdominal CT scans or ultrasonography, bone scintigraphy, and cranial CT scans or magnetic resonance imaging (MRI). All patients were followed up for more than 5 years. Each included patient was restaged based on the 8th edition of lung cancer staging classification (13).
Histological evaluation
All formalin-fixed, paraffin-embedded (FFPE) tissue specimens were recut and stained with hematoxylin and eosin (HE). Two pathologists concurrently re-evaluated the slides using a multiheaded microscope, and discrepancies were resolved through discussion. Histological subtypes of lung adenocarcinoma were classified on the basis of the International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society multidisciplinary classification of lung adenocarcinoma (1). Each histological subtype component was recorded in 5% increments. Based on the presence or absence of an MP/S growth pattern (Figure S1A), the study population was split into two groups: (I) patients with MP/S components and (II) patients without.
Image acquisition and tumor segmentation
Thin-section CT scans of the lungs were obtained at full inspiration by the SOMATOM Definition AS scanner (64×0.625 mm detector, 1.0 pitch; Siemens, Germany) or a Brilliance 40 scanner (40×0.625 mm detector configuration, 0.4 pitch, Philips, The Netherlands) with 120 KVp of tube energy and 200 mAS of effective dose. A medium sharp reconstruction algorithm was utilized for image reconstruction with a section thickness of 1 mm and a 0.7-mm increment. All CT scans were performed without contrast medium. The identified CT scans were downloaded from the Picture Archiving and Communication Systems (PACS) (Figure S1B).
We used the open-source platform (3D-slicer, v4.9.0, www.slicer.org) to achieve tumor segmentation via the “segment editor” model. Tumors were delineated on the CT images in horizontal, sagittal, and coronal planes using a semi-automatic segmentation based on “level tracing” and “smoothing”. A radiologist and a thoracic surgeon then reviewed all tumor segmentations in consensus, and any discrepancies were resolved by additional correction.
Radiomics feature extraction and selection
“PyRadiomics”, an open-source package for standardizing the extraction of radiomics data (https://github.com/Radiomics/pyradiomics), was used to extract 90 radiomics features from the nonfiltered segmented tumor regions (ROIs). The extracted features can be classified into five categories: (I) shape (n=14); (II) intensity (n=19); (III) gray level co-occurrence matrix (GLCM; n=25); (IV) gray level run length matrix (GLRLM; n=16); and (V) gray level size zone matrix (GLSZM; n=16). Shape features quantified the three-dimensional (3D) geometry of the tumor, such as sphericity and surface area. Intensity features, also called first-order statistical features, are derived from the histogram of the tumor voxel, and can include the 90th percentile and standard deviation. GLCM, GLRLM, and GLSZM features characterize tumor texture and reflect the intratumor heterogeneity via an analysis of the spatial patterns of the tumor voxel. A detailed description and computing algorithms of the radiomics features are given in the Appendices (Appendix 1). Each radiomics feature was compared between the two subgroups, and the corresponding P value was calculated to test the significance. Multicollinearity of the radiomics features was visualized through a correlation heatmap. To reduce the collinearity of the extracted features and remove redundant information to avoid the overfitting issue, the most significant feature was selected from each category by identifying the feature with the lowest P value within each category before modelling (14).
Model building and validation
Based on the selected radiomics features, four popular machine learning methods were utilized to build the predictive models. These methods included the generalized linear model (GLM), Naïve Bayes, random forest, and support vector machine (SVM). In order to evaluate the model’s stability, a bootstrap resampling technique was used, and 100 random samples were iteratively selected with replacement from the original dataset. In each iteration, the model was constructed on the selected samples and validated using the remaining patients in the original dataset. The discrimination ability of model was assessed with the area under the curve (AUC) value. In addition, the clinical usefulness of these models was measured and compared by a decision curve analysis.
Statistical analysis
Statistical analyses were performed in R platform (R version 3.4.2). R package “ComplexHeatmap” was used for unsupervised cluster analysis. Package “e1071” was used for the implementation of Naïve Bayes and SVM classifiers, while the package “randomForest” was applied to develop the random forest model, both using the default parameter tuning with caret interface. A GLM via penalized maximum likelihood was fitted using the “glm” function with default parameters. Continuous and categorical variables were analyzed through the student’s t and chi-squared tests, respectively. Variables with a P value <0.1 in univariate analysis were entered into the multivariate logistic regression in a forward stepwise fashion. A Kaplan-Meier analysis was used to evaluate the survival outcomes of different groups, and differences among the survival curves were evaluated by a log-rank test. A two-sided P value <0.05 was considered statistically significant.
Results
Baseline characteristics and survival analysis
From January 2011 to December 2013, a primary cohort consisting of 268 patients with invasive lung adenocarcinoma at Shanghai Pulmonary Hospital was included in this study. The median age was 61 years (range, 30–87 years). Most of the patients were female (56%) and had no smoking history (79.1%). Furthermore, 10.1% (27/268) of patients had lymph node metastasis, among whom 20 were N1 positive, 17 were N2 positive, and 10 had both N1 and N2 metastasis. With regards to histological subtypes, 77 adenocarcinomas (28.7%) were revealed as having MP components, among which only 9 were MP-predominant. In terms of solid growth pattern, 33 adenocarcinomas contained solid components, and 25 of them were solid-predominant. Overall, 36.6% (98/268) of lung adenocarcinoma patients had an MP/S component with 12 presenting both components. The validation cohort consisted of 193 patients, and, with the exception of slightly larger lung tumors (mean: 2.47 versus 2.17 cm; P=0.003), there were no significant differences in baseline characteristics compared to that of patients in the primary cohort (Table 1).
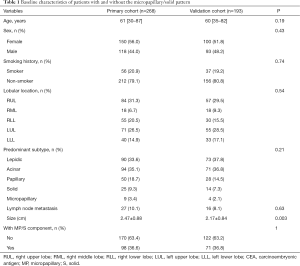
Full table
Clinical significance of the MP/S component
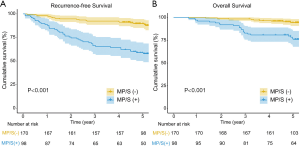
In univariate analysis, male patients with elevated CEAs and larger tumor sizes were more likely to be MP/S positive (Table S1). Among these three clinical factors, multivariate analysis demonstrated that sex was the only significant predictor of having an MP/S component. The presence of an MP/S component significantly correlated with tumor aggressive characteristics, including lymph node metastasis (18.4% versus 5.3%; P=0.001) and visceral pleural invasion (68.4% versus 55.3%; P=0.011). Overall and recurrence-free survival (OS and RFS, respectively) were compared between patients with MP/S components and those without. Log-rank tests revealed that the presence of an MP/S component was significantly associated with decreased OS and RFS in both the training (Figure 2, both P<0.001) and the validation (Figure S2, P<0.001 and P=0.016) cohort. Even for the patients without lymph node metastasis, the prognostic effect of an MP/S growth pattern still remained significant (Figure S3).
Radiomics analysis and feature selection
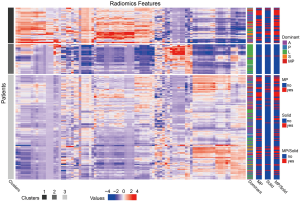
Unsupervised cluster analysis demonstrated that the radiomics feature was significantly associated with histological subtypes. As shown in Figure 3, three clusters of patients with similar radiomics pattern were significantly different in terms of predominant histological, MP-containing, and solid-containing subtypes, and the percentages of lung adenocarcinomas with an MP/S component were 62.5%, 1.9%, and 39.0%, respectively (P<0.001).
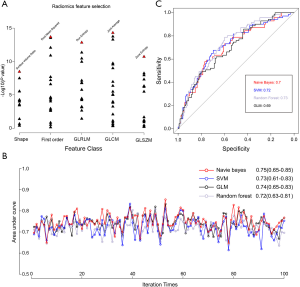
The correlation heatmap (Figure S4) revealed that multicollinearity was present among these radiomics features. By identifying the features with the lowest p values within each radiomics category, the five radiomics features contributing the most to the diagnosis of MP/S-containing lung adenocarcinomas were selected to compensate for this multicollinearity problem. As shown in Figure 4A, by calculating the negative log10 of the p value, the five radiomics features (surface volume ratio, root mean squared, run entropy, joint average, and zone entropy) were top-ranked in the y-axis. The differences in these five radiomics features between patients with MP/S components and those without are summarized in Table 2. In an unsupervised cluster analysis of the five selected radiomics features, the percentage of patients with MP/S components was significantly different among the formed clusters in both the primary and external cohorts (Figure S5).

Full table
Model building and validation
p>For the predictive model built by x, the probability of having an MP/S component can be simply expressed by the following equation: 1/[1+ exp (−0.0057× root mean squared − 1.93× zone entropy +25.4)]. The four machine leaning algorithms demonstrated similar model accuracy, with the Naïve Bayes model demonstrating the highest AUC value of 0.75 (95% CI: 0.65–0.85). The AUC values of each model in each iteration were all >0.6, which indicates good reliability of the selected radiomics features (Figure 4B). In the external validation, the AUC values were 0.70 (95% CI: 0.62–0.79), 0.72 (95% CI: 0.63–0.81), 0.73 (95% CI: 0.64–0.82), and 0.69 (95% CI: 0.61–0.77) for Naïve Bayes, SVM, random forest, and GLM, respectively (Figure 4C). The decision curve analysis indicated that there might be comparable net benefits among four proposed models (Figure S6).Discussion
Given that the MP and S subtypes present as strong predictors for tumor recurrence and worse survival of lung adenocarcinoma, we can confidently assume that predicting the presence of an MP/S component could be vital for improving the prognosis of lung adenocarcinoma (5,15). In this study, we confirmed that patients with an MP/S component had a higher rate of lymph node metastasis and worse prognoses than those without. In addition, we demonstrated that four machine learning models based on five optimal radiomics features could achieve similar and relatively accurate MP/S prediction performance (highest AUC value of 0.75; 95% CI: 0.65–0.85), which confirms the good reliability of radiomics features.
Overwhelming evidence shows that the MP and solid subtypes present as strong predictors for worse prognosis, and the prognostic effect remains even with proportions <5% [5,15]. Our results are consistent with these studies, which demonstrate that the presence of an MP/S growth pattern significantly correlated with lymph node metastasis (P=0.001) and visceral pleural invasion (P=0.011), as well as decreased OS and RFS. Furthermore, several reports (16-18) have also demonstrated that the MP pattern is positively associated with the EGFR mutation and anaplastic lymphoma kinase (ALK) fusion. It was also found that more than 85% of MP-predominant adenocarcinomas harbor EGFR driver mutations, and the MP growth pattern appears to be most likely correlated with a heterogeneous EGFR mutation, which acts as a mechanism of acquired resistance to EGFR tyrosine kinas inhibitors (TKIs) (18,19). Solid subtypes have otherwise been correlated with a high rate of KRAS mutation (20). Therefore, it is highly valuable to predict the presence of the MP/S growth pattern in helping to create personalized treatment and surveillance strategies for patients with lung adenocarcinoma.
Radiomics features can potentially decode the medical images to completely and non-invasively capture tumor phenotypic characteristics quantitatively, and has demonstrated promising predictive results for tumor invasiveness assessment (11), recurrence-free or overall survival (21,22), and treatment response (23). Regarding the histological growth pattern prediction, Park et al. (24) demonstrated that radiomics could differentiate three categories of the predominant subtypes in adenocarcinoma with favorable performance. However, the strong impact of MP/S component on poor prognosis needs to more accurately identify its presence, even those with a small portion. Song et al. (6) reported that a multivariable logistic model could identify two radiomics features, including the lower value for the minimum of whole pixel values and the lower value for the variance of positive pixel values, as imaging predictors for tumors with an MP component >5%. The unsupervised cluster analysis of our study also confirmed that the radiomics features are significantly associated with histological subtypes. However, the problem of multicollinearity leading to predictive model overfitting cannot be ignored. Jiang et al. (25) found that the random forest model built with reliable radiomics features (intraclass correlation coefficients ≥0.75) could obtain a good predictive performance in spreading through the air space in lung adenocarcinoma. Traverso et al. (26) proposed that machine learning–based radiomics methods may be useful for robust predictive model development. Thus in our study, after the pairwise comparison was performed to identify the most significant and predictive radiomics features from each category with the lowest P value, four popular machine learning methods were utilized to build the predictive models and achieve similar and stable performance in both internal and external validation. Our results indicated that these machine-learning methods turned out reasonably stable against data perturbation and hence they could be preferred for radiomics-based modeling. In addition, the five selected features were demonstrated to be highly reliable and could be utilized even when based on the CT images from different institutions, at different time points, and with different algorithms used for model construction.
This study also had some limitations which should be noted. First, both the primary and external validation cohorts were predominantly composed of female non-smoking patients with a relatively small sample size. Due to the imbalanced data seen in this study, a larger multicenter study would be needed for further validate the reproducibility and generalizability of our findings. Second, during radiomics feature extraction, the effects of CT scanner variability and inconsistent acquisition parameters were not considered. Further studies on radiomics should clarify the influence of the CT scanning setting on radiomics features, such as the application of the contrast medium. Lastly, the performance of the existed machine learning model for predicting the MP/S growth pattern was moderate and might be not yet suitable for clinical practice. The application of other novel ML-based predictive models or radiomics combined with deep learning in MP/S prediction may be worthwhile in future research.
In conclusion, radiomics features as applied to machine learning classification significantly correlate with histological subtypes of lung adenocarcinoma and seems to present a non-invasive, economical approach for MP/S growth pattern prediction.
Acknowledgments
The authors appreciate the academic support from AME Lung Cancer Collaborative Group. Financial supports from Shanghai Health Commission (2019SY072 & 2018ZHYL0102), Shanghai Pulmonary Hospital Research Fund (FK18001 & FKGG1805), Clinical Research Foundation of Shanghai Pulmonary Hospital (FK1944), Medicine and Public Health Scientific Projects in Zhejiang Province (2020KY270), and Huamei Key Research Foundation (2019HMZD05) were appreciated.
Funding: This study was supported by Shanghai Health Commission (2019SY072 & 2018ZHYL0102), Shanghai Pulmonary Hospital Research Fund (FK18001 & FKGG1805), Clinical Research Foundation of Shanghai Pulmonary Hospital (FK1944), Medicine and Public Health Scientific Projects in Zhejiang Province (2020KY270), and Huamei Key Research Foundation (2019HMZD05).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-21-44
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-21-44
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-21-44). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The waiver of informed consent was approved by the institutional review board (IRB) of each participating hospital, including Shanghai Pulmonary Hospital, Ningbo No. 2 Hospital, and the Affiliated Hospital of Zunyi Medical College. The study was performed in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Classification of lung adenocarcinoma. J Thorac Oncol 2011;6:244-85. [Crossref] [PubMed]
- Murakami S, Ito H, Tsubokawa N, et al. Prognostic value of the new IASLC/ATS/ERS classification of clinical stage IA lung adenocarcinoma. Lung Cancer 2015;90:199-204. [Crossref] [PubMed]
- Yoshizawa A, Sumiyoshi S, Sonobe M, et al. Validation of the IASLC/ATS/ERS lung adenocarcinoma classification for prognosis and association with EGFR and KRAS gene mutations: analysis of 440 Japanese patients. J Thorac Oncol 2013;8:52-61. [Crossref] [PubMed]
- Hung JJ, Jeng WJ, Chou TY, et al. Prognostic value of the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society lung adenocarcinoma classification on death and recurrence in completely resected stage I lung adenocarcinoma. Ann Surg 2013;258:1079-86. [Crossref] [PubMed]
- Cha MJ, Lee HY, Lee KS, et al. Micropapillary and solid subtypes of invasive lung adenocarcinoma: clinical predictors of histopathology and outcome. J Thorac Cardiovasc Surg 2014;147:921-928.e2. [Crossref] [PubMed]
- Song SH, Park H, Lee G, et al. Imaging Phenotyping Using Radiomics to Predict Micropapillary Pattern within Lung Adenocarcinoma. J Thorac Oncol 2017;12:624-32. [Crossref] [PubMed]
- Nitadori J, Bograd AJ, Kadota K, et al. Impact of micropapillary histologic subtype in selecting limited resection vs lobectomy for lung adenocarcinoma of 2cm or smaller. J Natl Cancer Inst 2013;105:1212-20. [Crossref] [PubMed]
- Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. [Crossref] [PubMed]
- Hawkins S, Wang H, Liu Y, et al. Predicting Malignant Nodules from Screening CT Scans. J Thorac Oncol 2016;11:2120-8. [Crossref] [PubMed]
- Zhong Y, Yuan M, Zhang T, et al. Radiomics Approach to Prediction of Occult Mediastinal Lymph Node Metastasis of Lung Adenocarcinoma. AJR Am J Roentgenol 2018;211:109-13. [Crossref] [PubMed]
- She Y, Zhang L, Zhu H, et al. The predictive value of CT-based radiomics in differentiating indolent from invasive lung adenocarcinoma in patients with pulmonary nodules. Eur Radiol 2018;28:5121-8. [Crossref] [PubMed]
- Parmar C, Grossmann P, Bussink J, et al. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci Rep 2015;5:13087. [Crossref] [PubMed]
- Detterbeck FC, Boffa DJ, Kim AW, et al. The Eighth Edition Lung Cancer Stage Classification. Chest 2017;151:193-203.
- Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. Erratum in: Nat Commun 2014;5:4644. [Crossref] [PubMed]
- Lee G, Lee HY, Jeong JY, et al. Clinical impact of minimal micropapillary pattern in invasive lung adenocarcinoma: prognostic significance and survival outcomes. Am J Surg Pathol 2015;39:660-6. [Crossref] [PubMed]
- Cai YR, Dong YJ, Wu HB, et al. Micropapillary: A component more likely to harbour heterogeneous EGFR mutations in lung adenocarcinomas. Sci Rep 2016;6:23755. [Crossref] [PubMed]
- Cai W, Lin D, Wu C, et al. Intratumoral Heterogeneity of ALK-Rearranged and ALK/EGFR Coaltered Lung Adenocarcinoma. J Clin Oncol 2015;33:3701-9. [Crossref] [PubMed]
- Ciompi F, Chung K, van Riel SJ, et al. Towards automatic pulmonary nodule management in lung cancer screening with deep learning. Sci Rep 2017;7:46479. [Crossref] [PubMed]
- Remon J, Majem M. EGFR mutation heterogeneity and mixed response to EGFR tyrosine kinase inhibitors of non small cell lung cancer: a clue to overcoming resistance. Transl Lung Cancer Res 2013;2:445-8. [PubMed]
- Zhang Y, Li J, Wang R, et al. The prognostic and predictive value of solid subtype in invasive lung adenocarcinoma. Sci Rep 2014;4:7163. [Crossref] [PubMed]
- Wang T, Deng J, She Y, et al. Radiomics Signature Predicts the Recurrence-Free Survival in Stage I Non-Small Cell Lung Cancer. Ann Thorac Surg 2020;109:1741-9. [Crossref] [PubMed]
- Yang L, Yang J, Zhou X, et al. Development of a radiomics nomogram based on the 2D and 3D CT features to predict the survival of non-small cell lung cancer patients. Eur Radiol 2019;29:2196-206. [Crossref] [PubMed]
- Coroller TP, Agrawal V, Huynh E, et al. Radiomic-Based Pathological Response Prediction from Primary Tumors and Lymph Nodes in NSCLC. J Thorac Oncol 2017;12:467-76. [Crossref] [PubMed]
- Park S, Lee SM, Noh HN, et al. Differentiation of predominant subtypes of lung adenocarcinoma using a quantitative radiomics approach on CT. Eur Radiol 2020;30:4883-92. [Crossref] [PubMed]
- Jiang C, Luo Y, Yuan J, et al. CT-based radiomics and machine learning to predict spread through air space in lung adenocarcinoma. Eur Radiol 2020;30:4050-7. [Crossref] [PubMed]
- Traverso A, Kazmierski M, Zhovannik I, et al. Machine learning helps identifying volume-confounding effects in radiomics. Phys Med 2020;71:24-30. [Crossref] [PubMed]


