
Comprehensive molecular analysis of genomic profiles and PD-L1 expression in lung adenocarcinoma with a high-grade fetal adenocarcinoma component
Introduction
Lung cancer is a leading cause of cancer-related mortality globally. Significant advancements in lung cancer treatment are based on the identification of the histologic subtypes and targeted agents, which are important factors that guide treatment-related decisions. Lung adenocarcinoma (LUAD) is the most common type of lung cancer, and invasive adenocarcinoma is classified into major histological patterns including lepidic, acinar, papillary, solid, and micropapillary, and variants such as mucinous, colloid, enteric, and fetal (1,2). Fetal adenocarcinomas are further divided into low-grade fetal adenocarcinomas (L-FLACs), and high-grade fetal adenocarcinomas (H-FLACs) (2,3). The association between prognosis and major histological patterns has been previously reported and indicate an association between solid/micropapillary patterns and poor prognosis (4-8). Few studies have focused on the prognosis of rare variants. We recently examined the prognostic influence of an H-FLAC component and revealed that LUADs with H-FLAC components had a poor prognosis comparable to that of micropapillary adenocarcinoma (9).
L-FLACs and H-FLACs display characteristic glandular structures resembling the airway epithelium during the pseudoglandular stage of fetal lung development and exhibit different clinicopathological and biological behaviors. L-FLACs reportedly constitute 0.3% of primary LUADs, usually appearing in the early stages in young-to-middle-aged women with a history of smoking, and display histological features including mild nuclear atypia and morula formation (3,10,11). H-FLACs constitute 0.5–1.4% of primary LUADs, frequently develop at advanced stages in elderly men with a history of heavy smoking, and display histological features including prominent nuclear atypia and the absence of morula formation (3,9,11-13). L-FLACs exhibit aberrant nuclear β-catenin localization with frequent CTNNB1 (β-catenin gene) mutations, whereas most H-FLACs do not display β-catenin aberrations (9,11-15). H-FLACs are therefore speculated to originate through a tumorigenesis process distinct from that of L-FLACs. However, the tumorigenesis process of H-FLACs remains unclear as only few systematic studies have focused on their genetic and molecular abnormalities, which may be considered as candidate molecular therapeutic targets.
Therefore, in this study, we performed whole-exome sequencing (WES) of 16 lung cancer cases with an H-FLAC component, to elucidate the genetic features of H-FLAC. Furthermore, based on the profile obtained from genetic alterations, an immunohistochemical (IHC) analysis of programmed death ligand-1 (PD-L1) expression was performed to predict the efficacy of immune checkpoint inhibitors for lung cancers with an H-FLAC component. To our knowledge, this is the first study to comprehensively assess H-FLAC. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-1158).
Methods
Case selection and clinicopathological evaluation
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the Kanagawa Cancer Center (approval no. H28-Res60) and individual consent for this retrospective analysis was waived. Sixteen samples of surgically resected primary lung cancers with an H-FLAC component present in various proportions (at least 5% of the total tumor volume) were obtained. All resected tissues were fixed in 10% formalin, embedded in paraffin, and stained with hematoxylin and eosin (HE). All cases were previously reported, and H-FLAC was histologically defined as previously described (9). The proportion of the H-FLAC component was evaluated as a percentage of the total tumor volume in HE-stained sections. Additionally, other coexisting components were assessed and classified in accordance with the World Health Organization classification of lung tumors (2). Clinical data including patient age, sex, smoking history, and tumor size and stage were retrieved from the Thoracic Surgical Database and medical records. Although the Union for International Cancer Control (UICC) published the 8th edition of the UICC TNM classification of malignant tumors in 2017 (16), the lung cancer stages in the study were evaluated in accordance with the 7th edition of the UICC TNM classification due to the difficulty in retrospective measurement of accurate invasive sizes of old lung cancer cases (17).
IHC analysis
IHC staining was performed for formalin-fixed paraffin-embedded (FFPE) tissue sections using an automated immunostainer (Histostainer 48A; Nichirei, Tokyo, Japan) with the following primary antibodies: mouse monoclonal anti-thyroid transcription factor-1 (TTF-1) (Agilent Cat# IR056, RRID:AB_2755006, dilution 1:200), anti-surfactant protein (SP)-A (IBL, Cat# 10375, RRID:AB_10679116, dilution 1:40), anti-SP-B (Santa Cruz Biotechnology, Cat# sc-17827, RRID:AB_2275110, dilution 1:400), anti-β-catenin (Leica, Cat# PA0088, RRID:AB_10555989, dilution 1:50), anti-p53 (Agilent Cat# M7001, RRID:AB_2206626, dilution 1:100), rabbit polyclonal anti-SP-C (Proteintech, Cat# 10774-1-AP, RRID:AB_2185497, dilution 1:200), and rabbit monoclonal anti-PD-L1 (Cell Signaling Technology, Cat# 13684, RRID:AB_2687655, dilution 1:200) antibodies. TTF-1, SP-A, SP-B, SP-C, β-catenin, and p53 expression was detected using the Histofine Simple Stain MAX-PO kit (Nichirei, Tokyo, Japan). PD-L1 expression was detected using Novolink Polymer Detection Systems (Leica Biosystems, Newcastle, United Kingdom). Appropriate positive and negative control tissues were used in each case. IHC staining for β-catenin, TTF-1, SP-A, SP-B, and SP-C was considered positive if these molecules were detected in >10% of H-FLAC cells. Upon IHC analysis of p53, staining of >50% of H-FLAC cells was considered as positive. The H-FLAC component was considered positive for PD-L1 expression if ≥1% PD-L1 staining was detected and was estimated in increments of 5% except for 1% positivity of H-FLAC cells.
WES
Multiple thinly sliced sections were prepared in succession from each FFPE tissue block, and the first section was stained with HE. Based on the results of HE staining, tissues from the H-FLAC component were manually obtained from the next 10 consecutive thinly sliced sections through macrodissection. DNA was extracted from the H-FLAC component, using the GeneRead DNA FFPE Kit (Qiagen, Hilden, Germany), in accordance with the manufacturer’s instructions, to reduce artefactual C>T|G>A transitions. Furthermore, DNA was extracted from non-neoplastic lung tissue in each case, using the same procedure to obtain the reference germline sequence. Double-stranded DNA was quantified using Qubit 2 (Thermo Fisher Scientific, Waltham, MA, USA). To assess DNA purity, the A260/A280 and A260/A230 optical density ratios were evaluated using NanoPhotometer (Implen, Munich, Germany).
In total, 200 ng of extracted DNA was used to generate panel libraries for each specimen. Libraries were amplified using SureSelect XT Target Enrichment System V6 (Agilent, Santa Clara, CA, USA) and sequenced using a Hiseq2500 system (Illumina, San Diego, CA, USA) with a standard 125-bp paired-end read protocol. We used the Genomon 2 DNA analysis pipeline (https://github.com/Genomon-Project) to detect somatic mutations and structural variations (SVs) for each case. Mutations such as single nucleotide variations (SNVs), short insertions and deletions (indels) with minimum depth ≥8, base quality ≥15, variant read ≥4, P<0.01 (Fisher’s exact test), and variant allele frequency ≥0.02 in the tumor, were selected as mutation candidates. Analysis of the candidates included predicting changes in amino acids, determining if the variants were previously reported as somatic changes and if the variants are potentially pathogenic. SNVs were further evaluated using the sequences obtained as queries against the Catalogue of Somatic Mutations in Cancer (COSMIC; https://cancer.sanger.ac.uk/cosmic) database, the 1000 Genomes Project (https://www.internationalgenome.org/) (frequency of variants <0.01), and dbSNP (NCBI, NIH; https://www.ncbi.nlm.nih.gov/snp/) [minor allele frequency (MAF) <0.01]. Furthermore, literature searches in combination with database searches were performed to exclude potential single nucleotide polymorphisms. SVs such as large insertions, deletions, tandem duplications, and inversions, with tumor allele frequency ≥0.07, depth thresh fold ≥10, control depth ≥10, removal of repeat sequence and inversion size threshold >1,000 in the tumor, were considered as SV candidates.
Analyses of tumor mutation burden (TMB), microsatellite instability (MSI), and copy number variations (CNVs)
To determine the TMB as numbers of somatic mutations per DNA megabase, SNVs and splice-site variants were considered. The MSI status was determined using MicrOSAtellite Instability Classifier (MOSAIC; https://github.com/ronaldhause/mosaic) (18). CNVs were identified as genomic regions with an abnormal copy number, using DNAcopy version 1.56.0. (https://bioconductor.org/packages/release/bioc/html/DNAcopy.html), an R/Bioconductor package.
Evaluation of mutation patterns and signatures
MutationalPatterns version 2.0.0. (https://bioconductor.org/packages/release/bioc/html/MutationalPatterns.html), an R/Bioconductor-based package developed by Blokzijl et al. (19), was used to characterize the broad range of patterns in base substitutions and mutational signatures identified via WES, to analyze the underlying mechanisms involved in the development and acquisition of the malignant characteristics of H-FLAC. Furthermore, data of the same analyses performed using a WES dataset comprising 514 LUADs obtained from The Cancer Genome Atlas (TCGA; https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) and the results were compared with those obtained for the H-FLACs in this study.
Statistical analysis
Mann-Whitney U test was performed to compare the TMB obtained from H-FLACs and the TMB obtained from LUADs in TCGA database. Cosine similarity was used to determine the relative contribution of the 30 COSMIC mutational signatures (Mutational Signatures version 2, https://cancer.sanger.ac.uk/cosmic/signatures_v2) in each sample.
Results
Clinicopathological features
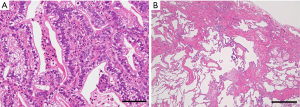
The clinicopathological findings for the 16 lung cancer cases with an H-FLAC component are summarized in Table 1. Lung cancers with an H-FLAC component were predominant among elderly men with a smoking history. Eight patients had stage I, five patients had stage II, and three patients had stage III disease. These cases displayed H-FLAC histology in various proportions: 5–9%, one case; 10–19%, two cases; 50–59%, one case; 60–69%, four cases; 70–79%, three cases; 80–89%, one case; 90–99%, two cases; and 100%, two cases. The H-FLAC component displayed complex glandular, cribriform, and papillary structures composed of columnar clear cells with pseudostratified atypical nuclei, resembling “piano keyboards” (Figure 1). Only two cases presented pure H-FLAC histology, whereas the others presented H-FLAC histology along with diverse components: conventional adenocarcinoma, 12 cases; invasive mucinous adenocarcinoma, three cases; enteric adenocarcinoma, one case; squamous cell carcinoma, one case; and sarcomatoid component, two cases. Pulmonary fibrosis with the usual interstitial pneumonia (UIP) pattern and severe emphysematous changes were observed in the background lung tissue in three (cases 4, 15, and 16) and two cases (cases 3 and 6), respectively.

Full table
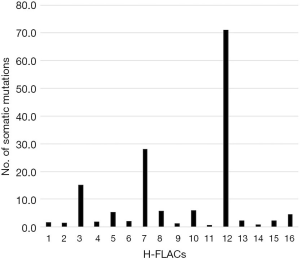
Somatic gene mutations identified via WES
The means of the reads obtained and the depth were 107,849,503 and 165.34, respectively. Each depth is indicated in Table S1. We found that 99.9% of the total reads were accurately aligned with the hg19 human genome reference sequence, thus indicating the accuracy and reliability of WES. The total number of somatic mutations satisfying the criterion set (P<0.01 and variant allele frequency ≥0.02) in each H-FLAC ranged from 26 to 2,402, and the TMB was 0.7–71.1 (median, 2.3; Figure 2). Through the same analysis, we obtained 12.5 as the TMB for the 514 TCGA LUADs, and the median TMB value of our cohort was significantly lower than that of the TCGA data (P<0.01). A high TMB of ≥10 mutations per Mb was observed in 3/16 cases (19%). All the nonsynonymous somatic mutations that led to peptide sequence alterations (including splice site mutations) identified in this study are included in table online (https://cdn.amegroups.cn/static/public/tlcr-20-1158-1.xlsx) along with the allele frequency.
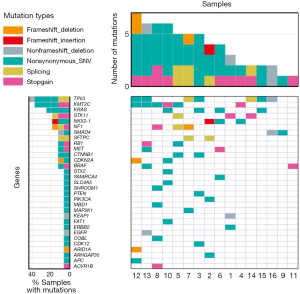
Nonsynonymous somatic mutations in H-FLACs were compared with a panel of 37 mutated genes in LUADs, as these genes displayed significant or near-significant mutation frequencies in 405 LUAD cases, and the data for this gene panel is available at TumorPortal (http://www.tumorportal.org/tumor_types?ttype=LUAD), hosted by the Broad Institute of MIT and Harvard (Cambridge, MA, USA). The most frequently mutated gene in our study was TP53 (7/16 cases, 44%), followed by KMT2C (also known as MLL3) (6/16 cases, 38%), KRAS (4/16 cases, 25%), NF1 (3/16 cases, 19%), and STK11 (3/16, 19%). SMAD4, RB1, MET, CTNNB1, CDKN2A, and BRAF mutations were reported in two cases each (2/16, 13% each). The other 14 mutated genes in LUAD, including EGFR and KEAP1, were observed in one case each. The H-FLACs shared 25 of the 37 (68%) genes mutated in LUAD; this may reflect the profile of common nonspecific LUADs.
We then analyzed the list of somatic hotspot mutations occurring in major cancers (20), as they may be the probable cancer driver genes even though they may not have been confirmed as driver genes through in vitro or in vivo analyses. Along with genes mutated in LUAD cases on TumorPortal, this approach identified PTEN, ACVR1B, COBL, KLF5, SHROOM1, and TPSD1; however, these mutations were observed in only one case each.
Furthermore, we analyzed the genes that are not well characterized as cancer-related genes but were repeatedly detected during the current WES analysis. We focused on two genes, NKX2-1 and SFTPC, involved in the pulmonary surfactant system. Three cases harbored NKX2-1 mutations, including two missense mutations and one frameshift deletion, and two cases harbored SFTPC mutations that were splice-site mutations. These results are summarized in Table S2 and schematically presented as the mutational landscape of H-FLAC in Figure 3. All KMT2C mutations were detected in lung cancers that contained ≥60% of the H-FLAC component. However, there was no appreciable difference in other gene mutations according to the proportion of H-FLAC component.
MSI status and CNVs
The MSI status was determined using the MicrOSAtellite Instability Classifier (MOSAIC), and a high MSI was not detected in any H-FLAC case. We analyzed genes harboring CNVs for the functional interpretation of large lists of genes, using DAVID (https://david.ncifcrf.gov/); however, none of the common CNVs were strongly associated with H-FLACs examined in this study (data not shown).
IHC analysis
The results of IHC analysis are displayed in Figure 4. The H-FLAC component displayed nuclear β-catenin localization in three cases, including two cases harboring a CTNNB1 mutation. One of the cases was a pure H-FLAC, and the other cases presented a combination with a lepidic adenocarcinoma component, which, in contrast, displayed membranous β-catenin staining. Diffuse p53 expression was detected in six cases, all presenting a TP53 mutation. Considering the results of WES and mutational signature analysis displayed below, TTF-1 and SPs including SP-A, SP-B, and SP-C were often not expressed in H-FLAC components.
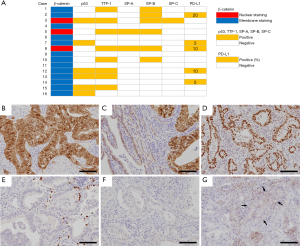
IHC analysis was performed for PD-L1 due to the significantly lower TMB in our H-FLAC cohort compared with the TCGA data of LUADs. Five cases displayed low PD-L1 expression (1–49% of H-FLAC cells), whereas the other cases were negative for PD-L1. In all cases wherein PD-L1 was not expressed in H-FLAC cells, it was not expressed in the other combined components as well.
Analyses of mutational patterns and mutational signatures
A plot of the six base substitution type spectra of all the 16 H-FLAC samples was obtained (Figure S1). Overall, the most common base change identified in H-FLACs was C>T; in contrast, the most common base change in the 514 cases of TCGA LUADs was C>A. Among the cases with the C>T base change, C>T was predominant in the non-CpG dinucleotide context, and this tendency was the same as that noted in the TCGA LUADs.
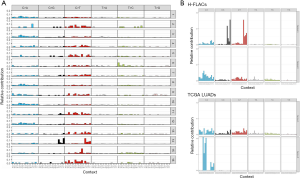
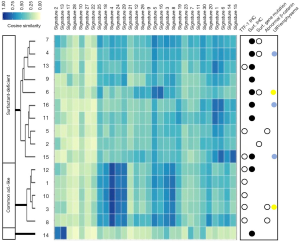
Thereafter, we obtained the mutation matrix of each of the 16 H-FLACs, which displayed a mutation pattern in terms of the relative contribution of the identified mutation numbers for all 96 trinucleotide changes (Figure 5A). Two common mutational signatures, designated as signatures A and B, were extracted de novo from the mutation matrices of 96 trinucleotide changes through non-negative matrix fractionation with the method provided in the MutationalPatterns package (Figure 5B). The same analysis was performed for the TCGA LUAD data, and the signatures extracted from them were markedly different (Figure 5B). The relative contribution of the well-known 30 COSMIC mutational signatures to the mutational profile of each H-FLAC was further evaluated by determining the cosine similarity through a MutationalPatterns package. H-FLACs were hierarchically clustered into three types in accordance with the cosine similarity to the COSMIC signatures. These clusters were well correlated with the immunostaining status of TTF-1 and the SPs SP-A, SP-B, and SP-C and to the status of surfactant gene mutations, designated as common adenocarcinoma-like (five cases), surfactant-deficient (ten cases), and signatures 2 and 13-related (one case) (Figure 6). All H-FLACs in the common adenocarcinoma-like cluster were positive for TTF-1, and all cases, except for one, displayed positive immunostaining for at least one of the three SPs. In contrast, H-FLACs in the surfactant-deficient cluster were negative for TTF-1 upon immunostaining, with three exceptions, and negative for the SPs, with two exceptions, and included all four cases of H-FLACs with mutations in the surfactant genes NKX2-1 or SFTPC. Additionally, all three cases with a mucinous adenocarcinoma component and one case with an enteric adenocarcinoma component were included in the surfactant-deficient cluster. Other coexistent histological components were not apparently associated with the mutational signature clusters. The common adenocarcinoma-like H-FLACs were strongly associated with signature 4 and, specifically, in comparison with the surfactant-deficient type, associated with signatures 3, 8, 24, and 29. The surfactant-deficient type was associated with signatures 1 and 6. The same analysis was performed for the 514 TCGA LUADs. Most TCGA LUADs were clustered into a group whose pattern in the cosine similarity heat map was quite similar to that of the common adenocarcinoma-like H-FLACs, revealing the appropriateness of our cluster nomenclature (Figure S2). Furthermore, a mutational signature pattern of the surfactant-deficient type of H-FLACs was similar to that of a minor cluster of TCGA LUADs.
Discussion
Molecular targeted therapy has been the standard systemic treatment method for patients with oncogene-driven advanced-stage non-small cell lung cancer (NSCLC); however, the currently targeted driver gene mutations for lung cancer were not detected in H-FLAC in this study, except for an EGFR mutation. We are currently analyzing RNA-sequencing data for RNAs extracted from the sample sets used for WES; currently no targetable gene fusion has been identified (data not shown). KRAS G12C inhibitors are being developed for treating cancers harboring this mutation (21,22); however, the G12C mutation was detected in only one case in this study. Six of the 16 cases (38%) harbored eight mutations in KTM2C, which was identified as the most frequently mutated gene in the present WES analysis. KTM2C is a key histone H3K4 monomethyltransferase along with KTM2D at enhancers, primarily implementing H3K4 monomethylation (23). Missense and nonsense somatic mutations in KTM2C have been identified in various malignancies including NSCLC, and KTM2C is inactivated through these mutations (24,25). According to the data in TumorPortal, 14% of LUADs harbored a somatic mutation in KTM2C, which was the third most frequently mutated gene along with KEAP1 (14%), following the most frequently mutated TP53 (51%) and KRAS (26%). The status of TP53 and KRAS mutations was similar between H-FLACs in this study and the LUADs from TumorPortal. Therefore, the high frequency of KTM2C mutations can be considered as a characteristic of H-FLACs. As H3K4 monomethylation is a histone mark of enhancer activation (24), perturbation of the epigenetic regulation of gene expression is potentially associated with the biological features of H-FLACs. Comparative genetic, epigenetic, and/or transcriptomic analyses of H-FLAC and non-H-FLAC components in the same tumors are required to further elucidate the characteristics of H-FLAC.
Although only one H-FLAC harbored an EGFR mutation, a markedly higher EGFR mutation frequency (46–64%) has been reported in conventional LUADs, especially among East Asian patients (7,26-29). The EGFR mutation rate in H-FLACs is reportedly low (6–20%), as also observed in our study (11-13). EGFR mutations are more prevalent among women and never-smokers among East Asian LUADs cases (26,27,29), whereas, H-FLAC reportedly occurs in elderly men with a smoking history (3,9,11-13). The gap in the EGFR mutation status potentially reflects differences in the etiological factors among lung cancer patients. The KRAS mutation associated with a smoking history and mucinous histology (7,30) was correspondingly detected in 25% of H-FLACs in this study. No significant differences between the cases displaying fetal-type histological morphology with or without driver mutations were noted.
Cellular genome integrity is always endogenously and exogenously challenged by DNA damage, and accumulated mutations can lead to diseases including cancer. Therefore, identification of the mutation signatures is important for determining the etiology of cancer (31). Based on the similarity of mutational patterns in H-FLACs with 30 well-established mutation signatures provided in COSMIC, we could classify H-FLACs into two major types based on the status of TTF-1 and surfactant expression, surfactant system gene mutations, and their association with UIP/severe emphysema. Although one-third of H-FLACs were classified as common adenocarcinoma-like H-FLACs, the remaining two-thirds could be considered as the surfactant-deficient type, with one exception that was strongly associated with signatures 2 and 13. Honda et al. performed WES for numerous LUAD cases and reported associations among mutations in pulmonary surfactant system genes (NKX2-1, SFTPA1, SFTPA2, SFTPB, and SFTPC), smoking, and a background of UIP (32). In this study, all three UIP cases and 1 of the 2 severe emphysema cases were included in the surfactant-deficient type of H-FLACs. Smoking or tobacco-chewing habits are associated with signatures 4 and 29 and were associated with the TTF-1-positive common adenocarcinoma-like type of H-FLACs in this study. Although clusters displaying mutational signature patterns similar to those of H-FLACs were observed in the cosine similarity heat map of TCGA LUADs, the proportions of clusters were markedly different between H-FLACs and TCGA LUADs. Because the association between mutational signatures and the predicted etiology remains unclear, precise clinicopathological studies with larger numbers of H-FLACs are required.
Owing to the heterogeneous mutational profiles and diverse combined non-fetal type histology, a previous study reported that H-FLAC is not a true entity but a morphological pattern (12), as revealed through the detection of various cancer-related mutations. Some mutational signature clusters identified in this H-FLAC cohort were shared with the LUAD datasets in TCGA. However, in rare cases, H-FLAC could occur in its pure form and can be confused with L-FLAC; therefore, H-FLAC should be recognized as a rare variant. Furthermore, regardless of the various genetic backgrounds, H-FLACs had characteristic clinicopathological features, unique fetal lung-like histology, and a potentially common oncogenic mechanism not detectable in this genomic study; hence, it should be considered as an independent entity of LUAD with fetal morphological appearance and high-grade behavior.
In this study, three cases presented aberrant nuclear β-catenin accumulation, and two of them harbored CTNNB1 hotspot mutations D32Y and D32V. All the samples in our previous study displayed membrane β-catenin localization (9); however, nuclear β-catenin localization was detected through IHC analysis in this study via another clone of the monoclonal antibody. No case of H-FLAC harboring a CTNNB1 mutation has been reported previously (11-13,15), unlike L-FLAC cases, which are characterized by CTNNB1 mutations and nuclear β-catenin localization (11,14,15). Therefore, H-FLAC is considered more similar to conventional LUAD than to L-FLAC. In this study, two cases with nuclear β-catenin localization were combined with the lepidic adenocarcinoma component, and the lepidic components did not display nuclear β-catenin localization. No L-FLAC component was observed in tumors in all cases. Therefore, it might be speculated that the H-FLAC component developed from a pre-existing lepidic component after the acquisition of the CTNNB1 mutation, although DNA sequencing of the lepidic components was not performed. However, Nakatani et al. reported that nuclear β-catenin localization was immunohistochemically detected in two of eight H-FLAC cases and that one of the two cases presented histologic features intermediate between L-FLAC and H-FLAC (15). L-FLAC may rarely progress to a high-grade form. Aberrant β-catenin localization is not considered sufficient for the differential diagnosis of L-FLAC and H-FLAC. The present results indicate that old age, prominent nuclear atypia, no morula formation, and the coexistence of a conventional adenocarcinoma component are important for distinguishing H-FLAC from L-FLAC.
In advanced NSCLC cases with high PD-L1 expression (≥50% of tumor cells), pembrolizumab, which is used for first-line immune checkpoint inhibitor monotherapy, reportedly improves progression-free and overall survival (33). In previous studies using the 22C3 PD-L1 IHC assay, which was approved as a companion diagnostic test for pembrolizumab by the Food and Drug Administration of the United States, positive PD-L1 staining (≥1% of tumor cells) was detected in 34–66% and high PD-L1 expression (≥50% of tumor cells) in 11–30% of NSCLC cases (33-37). In this study, positive PD-L1 staining (≥1% of H-FLAC cells) was observed in 5/16 (31%) cases; however, no sample displayed high PD-L1 expression (≥50% of H-FLAC cells). The E1L3N clone was used for the IHC analysis of PD-L1 in this study, owing to its reportedly higher sensitivity than the 22C3 clone (38). Only two a few studies have performed IHC analysis for PD-L1 in H-FLAC cases, and both studies reported cases negative for PD-L1 (39,40). Furthermore, the findings for MSI and TMB, which are surrogate markers for pembrolizumab application, did not indicate H-FLACs as a target for pembrolizumab treatment. Mutation signature 6, known to be associated with defective mismatch repair, and signature 1, known to be associated with an endogenous mutation process initiated by 5-methylcytosine deamination, are considered characteristics of the surfactant-deficient type. However, MSI analysis using MOSAIC in this study did not reveal any high MSI case, and the median TMB value in this cohort is significantly lower than that of TCGA LUADs. H-FLACs tend to display no/low PD-L1 expression and a comparatively low TMB/MSI, suggesting resistance to immunotherapy in these patients.
In conclusion, H-FLACs rarely harbor driver mutations that are currently targeted for lung cancers; however, they harbor highly frequent KMT2C mutations, implying the epigenetic regulation of the biological features of H-FLACs. Mutational signature analysis helped classify H-FLACs into two major types; however, further studies are required to clarify the underlying etiological factors. MSI, TMB, and the PD-L1 expression status suggest a poor response of H-FLACs to PD-L1 immune checkpoint therapy. As H-FLAC has a high malignant potential, further studies are required to identify candidate therapeutic target molecules.
Acknowledgments
We are grateful to the technical staff at Kanagawa Cancer Center for their expert technical assistance and the members of the Clinical Sequencing Team at Human Genome Center, Institute of Medical Science, The University of Tokyo, for their expert technical assistance and intellectual discussions. The super-computing resource was provided by the Human Genome Center, Institute of Medical Science, The University of Tokyo (
Funding: This work was supported in part by the Japan Society for the Promotion of Science (JP18K15111 to MS, JP16H06276 to YM).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1158
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-1158). Dr. SM reports other from Fujitsu Laboratories, Ltd., other from Liquid Mine, Ltd., outside the submitted work. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the Kanagawa Cancer Center (approval no. H28-Res60) and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/ European Respiratory Society International Multidisciplinary Classification of Lung Adenocarcinoma. J Thorac Oncol 2011;6:244-85. [Crossref] [PubMed]
- Travis WD, Brambilla E, Burke AP, et al. WHO classification of tumours of the lung, pleura, thymus and heart. Lyon: IARC Press, 2015.
- Nakatani Y, Kitamura H, Inayama Y, et al. Pulmonary adenocarcinomas of the fetal lung type: a clinicopathologic study indicating differences in histology, epidemiology, and natural history of low-grade and high-grade forms. Am J Surg Pathol 1998;22:399-411. [Crossref] [PubMed]
- Yoshizawa A, Motoi N, Riely GJ, et al. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol 2011;24:653-64. [Crossref] [PubMed]
- Warth A, Muley T, Meister M, et al. The novel histologic International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society classification system of lung adenocarcinoma is a stage-independent predictor of survival. J Clin Oncol 2012;30:1438-46. [Crossref] [PubMed]
- Tsuta K, Kawago M, Inoue E, et al. The utility of the proposed IASLC/ATS/ERS lung adenocarcinoma subtypes for disease prognosis and correlation of driver gene alterations. Lung Cancer 2013;81:371-6. [Crossref] [PubMed]
- Yoshizawa A, Sumiyoshi S, Sonobe M, et al. Validation of the IASLC/ATS/ERS lung adenocarcinoma classification for prognosis and association with EGFR and KRAS gene mutations: analysis of 440 Japanese patients. J Thorac Oncol 2013;8:52-61. [Crossref] [PubMed]
- Yanagawa N, Shiono S, Abiko M, et al. The Clinical Impact of Solid and Micropapillary Patterns in Resected Lung Adenocarcinoma. J Thorac Oncol 2016;11:1976-83. [Crossref] [PubMed]
- Suzuki M, Nakatani Y, Ito H, et al. Pulmonary adenocarcinoma with high-grade fetal adenocarcinoma component has a poor prognosis, comparable to that of micropapillary adenocarcinoma. Mod Pathol 2018;31:1404-17. [Crossref] [PubMed]
- Sato S, Koike T, Yamato Y, et al. Resected well-differentiated fetal pulmonary adenocarcinoma and summary of 25 cases reported in Japan. Jpn J Thorac Cardiovasc Surg 2006;54:539-42. [Crossref] [PubMed]
- Zhang J, Sun J, Liang XL, et al. Differences between low and high grade fetal adenocarcinoma of the lung: a clinicopathological and molecular study. J Thorac Dis 2017;9:2071-8. [Crossref] [PubMed]
- Morita S, Yoshida A, Goto A, et al. High-grade lung adenocarcinoma with fetal lung-like morphology: clinicopathologic, immunohistochemical, and molecular analyses of 17 cases. Am J Surg Pathol 2013;37:924-32. [Crossref] [PubMed]
- Suzuki M, Yazawa T, Ota S, et al. High-grade fetal adenocarcinoma of the lung is a tumour with a fetal phenotype that shows diverse differentiation, including high-grade neuroendocrine carcinoma: a clinicopathological, immunohistochemical and mutational study of 20 cases. Histopathology 2015;67:806-16. [Crossref] [PubMed]
- Proctor L, Folpe AL, Esper A, et al. Well-differentiated fetal adenocarcinoma of the lung: cytomorphologic features on fine-needle aspiration with emphasis on use of β-catenin as a useful diagnostic marker. Diagn Cytopathol 2007;35:39-42. [Crossref] [PubMed]
- Nakatani Y, Masudo K, Miyagi Y, et al. Aberrant nuclear localization and gene mutation of β-catenin in low-grade adenocarcinoma of fetal lung type: up-regulation of the Wnt signaling pathway may be a common denominator for the development of tumors that form morules. Mod Pathol 2002;15:617-24. [Crossref] [PubMed]
- James DB, Mary KG, Christian W (editors). TNM classification of malignant tumors. 8th edition. Hoboken: Wiley-Blackwell, 2017.
- Leslie HS, Mary KG, Christian W. TNM classification of malignant tumors. 7th edition. Hoboken: Wiley-Blackwell, 2009.
- Hause RJ, Pritchard CC, Shendure J, et al. Classification and characterization of microsatellite instability across 18 cancer types. Nat Med 2016;22:1342-50. [Crossref] [PubMed]
- Blokzijl F, Janssen R, van Boxtel R, et al. MutationalPatterns: comprehensive genome-wide analysis of mutational processes. Genome Med 2018;10:33. [Crossref] [PubMed]
- Chang MT, Asthana S, Gao SP, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol 2016;34:155-63. [Crossref] [PubMed]
- Canon J, Rex K, Saiki AY, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019;575:217-23. [Crossref] [PubMed]
- Hallin J, Engstrom LD, Hargis L, et al. The KRAS(G12C) Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov 2020;10:54-71. [Crossref] [PubMed]
- Hu D, Gao X, Morgan MA, et al. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Mol Cell Biol 2013;33:4745-54. [Crossref] [PubMed]
- Sze CC, Shilatifard A. MLL3/MLL4/COMPASS Family on Epigenetic Regulation of Enhancer Function and Cancer. Cold Spring Harb Perspect Med 2016;6:a026427 [Crossref] [PubMed]
- Liu P, Morrison C, Wang L, et al. Identification of somatic mutations in non-small cell lung carcinomas using whole-exome sequencing. Carcinogenesis 2012;33:1270-6. [Crossref] [PubMed]
- Song Z, Zhu H, Guo Z, et al. Correlation of EGFR mutation and predominant histologic subtype according to the new lung adenocarcinoma classification in Chinese patients. Med Oncol 2013;30:645. [Crossref] [PubMed]
- Kosaka T, Yatabe Y, Onozato R, et al. Prognostic implication of EGFR, KRAS, and TP53 gene mutations in a large cohort of Japanese patients with surgically treated lung adenocarcinoma. J Thorac Oncol 2009;4:22-9. [Crossref] [PubMed]
- Haneda H, Sasaki H, Lindeman N, et al. A correlation between EGFR gene mutation status and bronchioloalveolar carcinoma features in Japanese patients with adenocarcinoma. Jpn J Clin Oncol 2006;36:69-75. [Crossref] [PubMed]
- Mitsudomi T, Kosaka T, Endoh H, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol 2005;23:2513-20. [Crossref] [PubMed]
- Li H, Pan Y, Li Y, et al. Frequency of well-identified oncogenic driver mutations in lung adenocarcinoma of smokers varies with histological subtypes and graduated smoking dose. Lung Cancer 2013;79:8-13. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. Erratum in: Nature 2013 Oct 10;502(7470):258. Imielinsk, Marcin [corrected to Imielinski, Marcin]. [Crossref] [PubMed]
- Honda T, Sakashita H, Masai K, et al. Deleterious Pulmonary Surfactant System Gene Mutations in Lung Adenocarcinomas Associated With Usual Interstitial Pneumonia. JCO Precis Oncol 2018; [Crossref]
- Reck M, Rodriguez-Abreu D, Robinson AG, et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. [Crossref] [PubMed]
- Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018-28. [Crossref] [PubMed]
- Herbst RS, Baas P, Kim DW, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540-50. [Crossref] [PubMed]
- Chen Q, Fu YY, Yue QN, et al. Distribution of PD-L1 expression and its relationship with clinicopathological variables: an audit from 1071 cases of surgically resected non-small cell lung cancer. Int J Clin Exp Pathol 2019;12:774-86. [PubMed]
- Skov BG, Rorvig SB, Jensen THL, Skov T. The prevalence of programmed death ligand-1 (PD-L1) expression in non-small cell lung cancer in an unselected, consecutive population. Mod Pathol 2020;33:109-17. [Crossref] [PubMed]
- Munari E, Zamboni G, Lunardi G, et al. PD-L1 expression in non-small cell lung cancer: evaluation of the diagnostic accuracy of a laboratory-developed test using clone E1L3N in comparison with 22C3 and SP263 assays. Hum Pathol 2019;90:54-9. [Crossref] [PubMed]
- Kubouchi Y, Matsuoka Y, Araki K, et al. Pure high-grade fetal adenocarcinoma of the lung: a case report. Surg Case Rep 2018;4:70. [Crossref] [PubMed]
- Ishida H, Yasuda M, Nitanda H, et al. Pulmonary high-grade fetal adenocarcinoma associated with cystic airspace: A case report. Thorac Cancer 2020;11:1703-7. [Crossref] [PubMed]


