
Detection and quantification of EGFR T790M mutation in liquid biopsies by droplet digital PCR
Introduction
Lung cancer is the most commonly diagnosed cancer and remains the leading cause of cancer death (1). A significant improvement of progression-free survival has been achieved with receptor-tyrosine kinase inhibitors (TKIs) that target the epidermal growth factor receptor (EGFR) in patients with non-small cell lung cancer (NSCLC) harboring activating EGFR mutations (2-6).
Binding of the EGFR extracellular domain to its ligands triggers autophosphorylation at key tyrosine residues and activates several downstream signaling pathways. Certain mutations and/or amplification of the EGFR gene lead to constitutive activation of EGFR signaling and play an important role as oncogenic drivers in NSCLC. The prevalence of EGFR-activating mutations in a Caucasian population with lung adenocarcinoma is approximately 10–20%, and the most common (>90%) are small in-frame deletions in exon 19 and an amino acid substitution in exon 21 (L858R) (7-9). These alterations confer sensitivity to EGFR-TKI therapy, resulting in response rates up to 70% and median survival up to 24–30 months (10).
Despite initial responses, most patients with EGFR-mutant NSCLC and treated with EGFR-TKIs (such as gefitinib, erlotinib, and afatinib) will have disease progression within 9–14 months after starting the treatment (8,11). The major mechanism of acquired resistance to EGFR-TKIs is the occurrence of a secondary EGFR kinase domain mutation in exon 20, the T790M substitution, which accounts for about half of the cases (8,12,13). This mutation leads to an enhanced affinity for ATP, thus reducing the ability of ATP-competitive reversible EGFR tyrosine kinase inhibitors, including gefitinib and erlotinib, to bind to the tyrosine kinase domain of EGFR (14).
Recently, a third generation of EGFR-TKIs was developed that irreversibly block T790M mutant EGFR with maintained activity against the original exon 19del and L858R mutations (15). Thus, testing for the EGFR T790M mutation has become routine clinical practice in patients with NSCLC that become resistant to first- and second-generation EGFR-TKIs. Ideally, detection of this new mutation should be done in tumor tissue obtained by re-biopsy (9,16). However, many patients on progression develop lesions in inaccessible locations. Moreover, the poor performance status of the patients also makes re-biopsy difficult. It is estimated that up to 40% of relapsed NSCLC patients may be unable to provide a tumor tissue sample suitable for molecular analysis (17). For these patients it is acceptable to perform a liquid biopsy, which allows genotyping cell-free tumor DNA (cfDNA) present in the plasma and other body fluids (18).
Early comparisons between tumor tissue samples and liquid biopsy for determining EGFR mutation status concluded that analysis of cfDNA detected fewer EGFR mutation positive patients (19,20). However, subsequent studies using more sensitive assays such as the Inivata InVision™ (eTAm-Seq™) assay or the cobas EGFR Mutation Test, reported detection of the T790M mutation in plasma samples from 50% and 61% of the patients with NSCLC at disease progression after previous EGFR-TKI therapy (17,21).
Droplet digital PCR (ddPCR) is emerging as a very attractive option in the clinic to genotype cfDNA in liquid biopsies (18). This is a PCR method based on water-oil emulsion droplet technology. A cfDNA sample is fractionated into 20,000 droplets, PCR amplification of both the mutated and wild-type DNA molecules occurs in each individual droplet, and fluorescent specific probes are used to quantify the amplified molecules. Whether this approach has the required rigor to be used in the clinical setting remains debatable. A prospective validation study showed that plasma ddPCR detected EGFR T790M mutation with a sensitivity of 77%, supporting the use of this assay to direct clinical care (22). However, in a real-world setting, the practical sensitivity of the ddPCR assay may vary. Indeed, recent studies that analyzed plasma cfDNA by ddPCR reported values for the prevalence of the T790M mutation in patients with acquired resistance to EGFR-TKIs ranging between 30.4% (23) and 42.7% (24).
Here we present an optimized ddPCR strategy that was used to test for the presence of the resistance EGFR T790M mutation in plasma samples from 77 patients with NSCLC in progression, resulting in a positivity rate of 52%.
We present the following article in accordance with the STARD reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-1010).
Methods
Study population
This is a retrospective study including a total of 111 patients with NSCLC in progression after treatment with EGFR-tyrosine kinase inhibitors (EGFR-TKIs), who were tested for the presence of the resistance mutation T790M in exon 20 of the EGFR gene. Most patients were diagnosed with lung adenocarcinoma in TNM stage IV. The average age of the patients at progression was 67, ranging from 31 to 91 (Table 1). The EGFR-TKIs used as first- or second-line treatment are indicated in Table 1. All tests were performed at GenoMed, in Lisbon, Portugal, during the period from July 2015 to December 2019. The samples were obtained from the following hospitals in Portugal: Hospital Pulido Valente; Hospital de Santa Maria; Hospital Egas Moniz; Hospital dos Capuchos; Hospital Beatriz Ângelo; Hospital de Santo Espírito da Ilha Terceira; Hospital Professor Doutor Fernando Fonseca; Hospital da Luz; Hospital Garcia de Orta; CUF Descobertas; Unidade Local Saúde Norte Alentejano and Hospital de Cascais. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The Lisbon Academic Medical Center Ethics Committee approved the study (311/20). Because the study involves the secondary analysis of existing data collected during standard of care procedures, a waiver of informed consent was requested and approved. All the data were analyzed anonymously, thus protecting the privacy and personal identity information of participating individuals. Samples were collected for standard clinical care and were not used for any other purpose. The sample collection procedures used have minimal risks and no adverse events were reported. The performers of the DNA tests were informed that patients had a diagnosis of advanced NSCLC in progression after treatment with EGFR-TKIs. Additional patient-related information was provided by clinicians after DNA data analysis was completed.
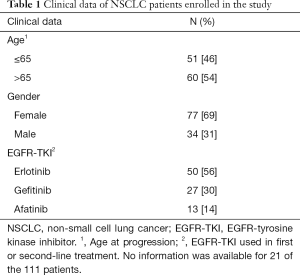
Full table
DNA analysis from FFPE samples
A trained pathologist made estimations of tumor cell content in formalin-fixed paraffin-embedded (FFPE) tissue samples resected from NSCLC patients. For macrodissection, the pathologist marked tumor areas on hematoxylin and eosin (H&E) stained tissue slides. Two to five serial FFPE sections of 10 μm thickness were then sliced from the selected areas. DNA was extracted using cobas® DNA Sample Preparation Kit, quantified using the NanoDrop® ND-1000 spectrophotometer (NanoDrop® Technologies), and analyzed by real-time PCR using cobas® EGFR Mutation Test. For Idylla™ EGFR Mutation Assay, sample preparation was done according to the manufacturer’s instructions.
Isolation of cfDNA from plasma
To extract cfDNA from plasma, ~10 mL blood samples were collected in EDTA or Cell-Free DNA BCT® (Streck) tubes. After centrifugation (900 g for EDTA tubes and 1,600 g for Streck tubes) for 10 minutes at room temperature, the supernatant was transferred into 2 mL microtubes, wasting about 5 mm of plasma in order to avoid buffy-coat disturbance. The samples were then centrifuged at 16,000 g for 10 additional minutes at room temperature to remove cell debris, thus reducing contamination with cellular DNA. The supernatants were collected into microtubes (2 or 5 mL), frozen, and stored at −80 °C. The cfDNA was isolated from 2 mL thawed plasma samples using QIAamp® MinElute® ccfDNA (QIAGEN), according to the manufacturer’s instructions. The cfDNA was eluted in 50 µL of ultra-clean water and quantified using the Qubit® 3.0 Fluorometer (Invitrogen, Life Technologies) with Qubit® dsDNA HS Assay kit (Invitrogen, Life Technologies). The purified cfDNA was stored at 4 °C for up to 16 hours or at −80 °C for longer periods.
T790M screening by droplet digital PCR (ddPCR)
To screen for the T790M mutation in plasma cfDNA, we used the QX200™ Droplet Digital PCR System (Bio-Rad, Hercules, CA, USA), and Bio-Rad assay primer/probe mixtures (dHsaCP2000019 and dHsaCP2000020). As reference for wild type and mutant EGFR, we used Horizon DNA standards HD709 and HD258 (Horizon Discovery Ltd., Cambridge, UK). The optimal annealing temperature was established at 59 °C. Altogether, the thermocycling conditions included an initial incubation at 95°C for 10 minutes, 47 cycles of 94 °C for 30 seconds and 59 °C for 1 minute, followed by an inactivation step at 98 °Cfor 10 minutes. At least two replicates were analyzed per sample, and the total amount of DNA loaded (considering the two replicates), was approximately 15 ng. No template control (NTC) was used to exclude PCR contamination. Amplification results were analyzed using QX200™ Droplet Reader and QuantaSoft™ software. Samples with three or more positive mutant droplets were considered positive, as recommended by the best practice guidelines for rare mutation detection (25). If one or two droplets were observed, the result was considered inconclusive and whenever possible a second sample was collected and tested.
Statistical analysis
The concentrations of target alleles were calculated using QuantaSoftTM software (Bio-Rad) based on Poisson distribution. Mutant cfDNA molecules were reported as number of copies per milliliter (mL) of plasma. The mutant allelic frequency was determined as the ratio of mutant droplets relative to the sum of mutant and wild type droplets.
Results
Quality assessment
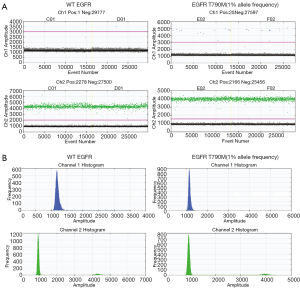
DNA reference standards for wild type and mutant EGFR confirmed the specificity of the ddPCR primers and probes (Figure 1). The wild type DNA exhibited high fluorescence signal in the hexachloro-6-carboxy-fluorescine (HEX) channel (channel 2, green) and no fluorescence signal in the fluorescein amidite (FAM) channel (channel 1, blue) (Figure 1A). In contrast, when the wild type and the mutant DNA were mixed at 1% allele frequency, fluorescence signal in the FAM channel (channel 1, blue) was detected (Figure 1B).
Next, we tested a series of 13 non-clinical liquid biopsy reference samples spanning different mutations and allele frequencies. These samples were provided by AstraZeneca and none of the laboratory technicians were informed about the genotype or mutant allelic frequencies at the time of execution of the tests. The results show that our ddPCR assay accurately detected the T790M allele in frequencies ranging from 5 to 0.5% (Table 2).
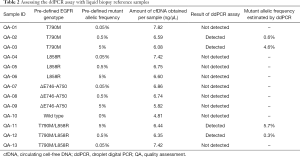
Full table
Finally, we applied the ddPCR assay to analyze DNA extracted from FFPE tissue biopsies from NSCLC patients. These DNA samples were previously genotyped in our laboratory by real-time PCR using cobas® EGFR Mutation Test. The ddPCR assay detected the mutation in all samples that previously tested positive for the T790M allele.
EGFR mutation testing in clinical plasma samples
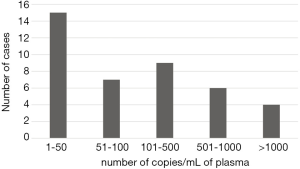
The ddPCR assay was used to screen plasma samples from 77 patients for the T790M mutation in EGFR. A total of 40 plasma samples were identified as positive for the T790M mutation, corresponding to 52%. The number of mutant molecules per mL of plasma ranged from 1 to 6,000 as depicted in Figure 2. A second sample was collected for 18 patientsthat initially tested negative (Table 3). As recommended by current guidelines (9,16), when the liquid biopsy was negative, re-biopsy was performed for analysis of tumor tissue. However, this was only possible in 12 cases, either because the patient status did not allow re-biopsy or the tumor was not accessible. We detected the T790M mutation in 2 of the 12 re-biopsies (17%) (Table 3). In the remaining cases, we analyzed either a second plasma sample (5 patients) or a sample of bronchoalveolar fluid (1 patient), and detected the T790M mutation in 3 of these 6 liquid biopsies (50%) (Table 3).
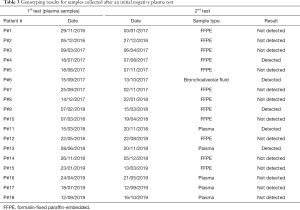
Full table
Finally, we analyzed the association of T790M detected in progression with EGFR activating mutations detected at diagnosis (Table 4). The majority (61%) of tumors positive for T790M had an EGFR del19 mutation at diagnosis, whereas the majority (65%) of tumors negative for T790M had either L858R or other EGFR mutations (Table 4).

Full table
Discussion
We implemented a liquid biopsy ddPCR assay that enabled the detection and quantification of the EGFR T790M mutation at cfDNA allele frequency as low as 0.5%. With a turnaround time of 3–5 business days, the assay was routinely used in clinical practice since it played an important role in deciding the next line of treatment in NSCLC patients that acquired resistance to first- or second-generation EGFR-TKIs.
We analyzed plasma samples from 77 patients and detected the EGFR T790M mutation in 52%. Although this was a retrospective study potentially influenced by bias associated with patient selection, our result is in good agreement with previous evidence indicating that about half of lung cancers that become resistant to EGFR TKIs acquire the EGFR T790M mutation (13). Most important, our results revealed that performing the ddPCR assay in a second liquid biopsy collected 1–8 months after a first negative plasma test increased by 50% the number of positive cases. Thus, the use of repeated ddPCR-based cfDNA genotyping may obviate tissue re-biopsy in cases that the tumor is not accessible or the patient has a poor functional status.
In our study, among tumors harboring the T790M mutation detected in either liquid biopsy or tumor tissue at disease progression, the majority (61%) had an EGFR del19 mutation at diagnosis. This is consistent with previous reports indicating that the T790M mutation is more frequent in patients with an EGFR exon 19 deletion mutation. In a cohort of 314 Japanese patients studied by re-biopsy, the T790M mutation was detected in 55.6% of cases with del19 mutation and in 43.0% of cases with the L858R mutation (26). In another study, the T790M mutation was present in 63% (26/41) of patients with EGFR exon 19 deletion and in 38% (12/32) of patients with L858R mutation (27). A more recent literature review confirmed that detection of the T790M mutation was more frequent in del19 mutated patients (53%) than in L858R mutated patients (36%) with acquired resistance to EGFR-TKIs (28). One possibility is that patients with the del19 mutation are more sensitive to TKIs, and therefore cells with the T790M mutation are more likely to be selected and enriched (28).
An important advantage of ddPCR-based assays is the ability to provide absolute quantification of DNA molecules. In our study cohort, the number of T790M mutant molecules per mL of plasma ranged from 1 to 6,000. Quantification of mutant molecules in plasma may be useful to monitor response to treatment. Indeed, a recent study reported that after 6 weeks of treatment with osimertinib, the T790M mutation load assessed by ddPCR decreased to very low level (23). However, whether the number of mutant molecules affects disease progression and response to therapy remains unknown. Clearly, future studies using quantitative diagnostic assays such as ddPCR are needed to maximize the benefits of precision therapy for advanced NSCLC.
Acknowledgments
We thank AstraZeneca for providing non-clinical control samples.
Funding: This work was supported by Fundação para a Ciência e Tecnologia (FCT)/Ministério da Ciência, Tecnologia e Ensino Superior - Fundos do Orçamento de Estado (UID/BIM/50005/2019), and FCT/FEDER/POR Lisboa 2020, Programa Operacional Regional de Lisboa, PORTUGAL 2020 (LISBOA-01-0145-FEDER-016394). C.S. was a recipient of a FCT fellowship (SFRH/BDE/110544/2015).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1010
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-1010
Peer Review File: Available at http://dx.doi.org/10.21037/tlcr-20-1010
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-1010). ACS reports receiving grants from AstraZeneca, as well as personal fees and non-financial support from AstraZeneca and Novartis. CS reports receiving personal fees from AstraZeneca. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of Centro Hospitalar Universitário Lisboa Norte and Lisbon Academic Medical Center (n. 311/20). Because the study involves the secondary analysis of existing data collected during standard of care procedures, a waiver of informed consent was requested and approved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or Carboplatin-Paclitaxel in Pulmonary Adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Yang JCH, Sequist LV, Geater SL, et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: A combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol 2015;16:830-8. [Crossref] [PubMed]
- Wu YL, Cheng Y, Zhou X, et al. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): a randomised, open-label, phase 3 trial. Lancet Oncol 2017;18:1454-66. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in untreated EGFR-Mutated advanced non-small-cell lung cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497-500. [Crossref] [PubMed]
- Stewart EL, Tan SZ, Liu G, et al. Known and putative mechanisms of resistance to EGFR targeted therapies in NSCLC patients with EGFR mutations-a review. Transl Lung Cancer Res 2015;4:67-81. [PubMed]
- Planchard D, Popat S, Kerr K, et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2018;29:iv192-237. [Crossref]
- Morgillo F, Della Corte CM, Fasano M, et al. Mechanisms of resistance to EGFR-targeted drugs: Lung cancer. ESMO Open 2016;1:e000060 [Crossref] [PubMed]
- Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015;21:560-2. [Crossref] [PubMed]
- Gazdar AF. Activating and resistance mutations of EGFR in non-small-cell lung cancer: role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 2009;28:S24-31. [Crossref] [PubMed]
- Sequist LV, Waltman BA, Dias-Santagata D, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 2011;3:75ra26 [Crossref] [PubMed]
- Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A 2008;105:2070-5. [Crossref] [PubMed]
- Murtuza A, Bulbul A, Shen JP, et al. Novel third-generation EGFR tyrosine kinase inhibitors and strategies to overcome therapeutic resistance in lung cancer. Cancer Res 2019;79:689-98. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med 2018;142:321-46. [Crossref] [PubMed]
- Jenkins S, Yang JCH, Ramalingam SS, et al. Plasma ctDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non–Small Cell Lung Cancer. J Thorac Oncol 2017;12:1061-70. [Crossref] [PubMed]
- Rolfo C, Cardona AF, Cristofanilli M, et al. Challenges and opportunities of cfDNA analysis implementation in clinical practice: Perspective of the International Society of Liquid Biopsy (ISLB). Crit Rev Oncol Hematol 2020;151:102978 [Crossref] [PubMed]
- Goto K, Ichinose Y, Ohe Y, et al. Epidermal growth factor receptor mutation status in circulating free DNA in serum: From IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol 2012;7:115-21. [Crossref] [PubMed]
- Mok T, Wu YL, Lee JS, et al. Detection and dynamic changes of EGFR mutations from circulating tumor DNA as a predictor of survival outcomes in NSCLC Patients treated with first-line intercalated erlotinib and chemotherapy. Clin Cancer Res 2015;21:3196-203. [Crossref] [PubMed]
- Remon J, Caramella C, Jovelet C, et al. Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA. Ann Oncol 2017;28:784-90. [Crossref] [PubMed]
- Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and kras mutations in advanced lung cancer. JAMA Oncol 2016;2:1014-22. [Crossref] [PubMed]
- Li C, Jia R, Liu H, et al. EGFR T790M detection and osimertinib treatment response evaluation by liquid biopsy in lung adenocarcinoma patients with acquired resistance to first generation EGFR tyrosine kinase inhibitors. Diagn Pathol 2018;13:49. [Crossref] [PubMed]
- Guo QM, Wang L, Yu WJ, et al. Detection of Plasma EGFR Mutations in NSCLC Patients with a Validated ddPCR Lung cfDNA Assay. J Cancer 2019;10:4341-9. [Crossref] [PubMed]
- Rare Mutation Detection Best Practices Guidelines [Internet]. 2015 Feb [cited 2020 May]; Available online: https://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_6628.pdf
- Nosaki K, Satouchi M, Kurata T, et al. Re-biopsy status among non-small cell lung cancer patients in Japan: A retrospective study. Lung Cancer 2016;101:1-8. [Crossref] [PubMed]
- Matsuo N, Azuma K, Sakai K, et al. Association of EGFR Exon 19 Deletion and EGFR-TKI Treatment Duration with Frequency of T790M Mutation in EGFR-Mutant Lung Cancer Patients. Sci Rep 2016;6:36458. [Crossref] [PubMed]
- Liang H, Pan Z, Wang W, et al. The alteration of T790M between 19 del and L858R in NSCLC in the course of EGFR-TKIs therapy: A literature-based pooled analysis. J Thorac Dis 2018;10:2311-20. [Crossref] [PubMed]


