
Increased detection of circulating tumor DNA by short fragment enrichment
Introduction
Although tumor tissue biopsy is currently considered the gold standard for the diagnosis and molecular characterization of tumors, it has several limitations. First, agnostic procedures eive procedures altnt. as several limitat changed with this "ire compresse con Plasil fl intrtissue biopsy is subjected to selection bias and may be inadequate to depict the whole molecular profile due to tumor heterogeneity. Moreover, it is rarely feasible in the clinical setting to obtain serial tissue biopsies from the same patient to track tumor genomic evolution over time (1,2). Compared with tissue biopsies, liquid biopsy is not limited by tumor heterogeneity and the problem of repeated sampling. Liquid biopsy thus has substantial potential as a tool for determining the genomic profile of patients with cancer, monitoring treatment responses, quantifying minimal residual disease, and assessing the emergence of therapy resistance (2-4). A range of tumor components can be isolated from the blood, with circulating tumor DNA (ctDNA) attracting considerable attention in the past 10 years due to its ability to reflect the mutation profile of tumor (5,6). Through the development of an array of techniques, such as large-scale parallel sequencing and ultra-deep sequencing (e.g., Duplex sequencing and integrated digital error suppression-enhanced CAPP-seq), the sensitivity and specificity of ctDNA assays have been improved significantly (7-9). The use of ctDNA analysis has begun the transition into clinical practice, and an increasing number of ctDNA tests for specific cancers have been approved by the major global regulatory bodies, including the Chinese National Medical Products Administration (NMPA), the U.S. Food and Drug Administration (FDA), and the European Medicines Agency (EMA).
Cell-free DNA (cfDNA) is released into the blood after cell necrosis or programmed death, so tissues with high metabolism tend to release more DNA, such as hematopoietic tissue, tumor tissue. cfDNA are also derived from a graft, a fetus or autonomously activated DNA. There is a peak at about 167 bp (corresponding to the length of nucleosome), and the other peaks have 10.5 bp cycle fluctuation, corresponding to the pitch of nuclear body core DNA.
ctDNA is part of the cfDNA pool. ctDNA is released from tumor cells and differs from the cfDNA derived from normal cells in some characteristics. One of the most significant difference is segment length. A number of studies have reported the increased integrity of tumor-derived plasma DNA, whereas others have found evidence suggesting that plasma DNA molecules released by tumors might be shorter (10-16). Mouliere et al. stated that this inconsistency across studies was due to the limitation in determining the specific sizes of tumor-derived DNA fragments, and that a more detailed characterization of changes in matched tumor-derived alterations and a broader understanding of potential biological differences are required, but lacking (17). Their research showed that mutant ctDNA was more fragmented than non-mutant cfDNA, and the size of ctDNA was approximately 20–40 bp shorter than that of nucleosomal DNA sizes, with enrichment occurring in segments 90–150 bp in size and 250–320 bp in size (17).
The difference in size between ctDNA with mutations and cfDNA without mutations might provide a new way to improve the sensitivity of ctDNA detection. Recently, several studies have confirmed this possibility mainly through the use of three methods (16-21). The first method is direct size-selection for shorter cfDNA or amplifying the library through agarose matrix separation and extraction by using an automated liquid handler (NIMBUS Select, Hamilton, Reno, NV, USA) or the PippinHT/Blue Pippin (Sage Bioscience, Beverly, MA, USA) (17,19). Selecting fragments between 90–150 bp has been found to improve the detection of tumor DNA with more than 2-fold enrichment in >95% of cases and more than 4-fold enrichment in 10% of cases (17). Another study showed that the variant alleles of short fractions (median insert size: ~142 bp) were on average two-fold enriched compared to the original cfDNA (median insert size: ~167 bp) (19). The second approach includes using biological information directly to acquire and analyze shorter reads or the ratio of short and long reads (17,18). The third method involves single-strand DNA (ssDNA) library preparation, which is particularly useful or managing degraded and fragmented DNA and improving library efficiency (20,21). One study compared ssDNA libraries with double-strand DNA (dsDNA) libraries and found that ssDNA libraries contained higher ctDNA, with the higher ctDNA content being associated with shorter insert size (20).
Here, we present a method for ssDNA library preparation with a large proportion of magnetic beads to enrich the shorter cfDNA. The purpose of this study was to determine if the approach could increase the ctDNA content and improve the sensitivity of ctDNA detection. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tlcr-21-180).
Methods
Patient and sample characteristics
We used a commercial cfDNA reference (cat. no. HD780, Multiplex I cfDNA Reference Standard Set, Horizon Discovery, Cambridge, UK) and 28 cfDNA samples from different cancer patients (22 non-small cell lung cancer, one esophageal squamous cell carcinoma, one cholangiocarcinoma, one liver cancer and one colorectal cancer patients), with different mutation (involved EGFR, KRAS and PIK3CA) frequencies ranging from 0.5% to 4.89%, as detected by next-generation sequencing (NGS) using NovoPM panel (Novogene Co., Ltd., China). Of the 28 samples, 2 were used to construct both a library with a large proportion of magnetic beads (the L-library) and a library with a regular proportion of magnetic beads (the R-library), while the other 26 were used to construct the L-library only, with 2 of them being used for the three-dimensional polymerase chain reaction (3D PCR) test. All patients provided informed consent, and the study was approved by institutional review boards at the Henan Cancer Hospital, China. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Plasmid construction and small fragment cfDNA simulation
Designed fragment used in this study were synthesized by Sangon Biotech (Shanghai, China) and contained specific mutations with a ~150 bp flanking wild-type sequences on both sides (the sequence information is shown in Table S1). Ten of these fragments were inserted into the T-vector pUC57. After verification, the plasmids were enriched for use.
The constructed plasmids were then fragmented using an S220 Focused-ultrasonicator (Covaris Inc., Woburn, MA, USA) with the following parameters: peak incident power, 175; duty factor 50%; cycles per burst, 200; and treatment time, 600 s. After fragmentation, the DNA was cleaned up with 3.0× magnetic beads.
The plasmid DNA was then diluted and spiked in the cell line (United States National Human Genome Cell Bank, NA10840) genomic DNA (gDNA), which was cultured using 1640 with 2 mM L-glutamine, 15% FBS and extracted by micrococcal nuclease (TaKaRa, cat. no. 5333) to mimic the cfDNA fragment size. The plasmid DNA was diluted at three different temperatures to produce different frequencies. Finally, libraries using a regular and a large proportion of magnetic beads were constructed with both a 20 ng and 5 ng input of total cfDNA.
Library preparation, target enrichment, and DNA sequencing
For each sample, different inputs of cfDNA were used to prepare ssDNA libraries (cat. no. 10096, Accel-Ngs® 1s Plus DNA Library Kit, Swift Biosciences, Ann Arbor, MI, USA) depending on the verified frequency and the remaining amount of cfDNA. For frequencies of <1.5%, 1.5–4, and >4%, the input of DNA was 10 ng, 5 ng, and 2 ng respectively, except for 1 sample (2 mutant sites) in which 2.55 ng was used due to the insufficient amount of the cfDNA (Table S2). The dsDNA library preparation method included end-repair, adapter ligation, and amplification. We purified these dsDNA libraries with a bead ratio of 1.0 (beads-to-sample ratio 1:1) using VAHTS DNA Clean Beads (cat. no. 411, Vazyme Biotech Co., Ltd., Nanjing, China). The ssDNA library preparation included denaturation, adaptase, extension, adaptor ligation, and amplification. At the points of post-extension, post-ligation and post-PCR cleanup, the ratio for conventional library construction was modified to ratios 1.8, 1.6, and 1.6, respectively. This modification was able to better recover the fragments longer than 40 bp.
A total of 500 ng of pre-libraries was employed for target enrichment through use of our of customized panel (IDT). Briefly, after the library adaptor was blocked and the DNA sequence was repeated, the library was denatured and hybridized with captured probes; after 16 hours of hybridization, the enriched library was pulled down using M270 Dynabead Streptavidin Beads (cat. no. 65306, Thermo Fisher Scientific, USA), and non-specific segments were washed away. Finally, the targeted DNA was enriched and amplified, and the PCR products were cleaned up. The concentration of the enriched library was measured by using the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific).
All cfDNA libraries were sequenced using 150 bp of paired-end runs on the NovaSeq 6000 system (Illumina Inc., San Diego, CA, USA).
Read mapping and single-nucleotide variant/insertion-deletion (SNV/INDEL) analysis
For sequencing the adapters, low quality bases were trimmed from raw sequencing reads using fastp software, and then 8 bases of the 3' end were trimmed. After adapter trimming and quality control, reads were mapped to the UCSC hg19 reference genome using BWA-MEM (version 0.7.17) under the default settings except that the minimum seed length was set to 32. Next, duplicates were marked with samblaster. Then, VarScan 2 was applied to all SNV/INDEL variants using the following settings: “—min-coverage 100—min-reads2 4—min-avg-qual 30—min-var-freq 0.005—P value 0.05—min-freq-for-hom 0.75—strand-filter 0—output-vcf 1—variants 1”. Finally, the VarScan 2 software tool, fpfilter, was used to decrease the false-positive rate. Of note, the BAM was further checked using Integrative Genomics Viewer (IGV) to remove potential artifacts.
Statistical analysis
All statistical analyses were performed by R3.6.3. We used the Wilcoxon rank-sum test for analyses that compared two groups and considered a P value of lower than 0.05 to be statistically significant.
Results
Increasing the size of short fragment cfDNA
It is acknowledged that different magnetic bead ratios can recover different sizes of DNA fragments. As a general rule, increasing the ratio of bead volume to sample volume will increase efficiency of binding smaller fragments, which means that using a greater bead ratio can recover even shorter fragments. The fragment distribution of tumor-derived cfDNA (ctDNA) is smaller than the cfDNA derived from healthy cells. We thus speculated that using a large proportion of magnetic beads might yield more tumor-derived cfDNA in the process of library construction. First, we detected the library fragment distribution using a cfDNA standard (Horizon Discovery) with large and regular proportions of magnetic beads in the process of ssDNA library construction (Swift Biosciences). The result showed that the ratio of fragments ranging from 90 to 150 bp in the library using a large proportion of magnetic beads (the L-library) was greater than that in the regular library (the R-library) (48.9% in the L-library vs. 14.3% in the R-library; Figure S1). We then used two clinical samples to determine the real-world similarity. As shown in Figure 1A,B, the inserted DNA fragments of clinical cfDNA in the L-library were shorter than those in the R-library; meanwhile, the ratios of fragments ranging from 91 to 150 bp in the L-library were 17.9% and 21.7%, while the ratios in the R-library were 7.2% and 8.5%. More specifically, the median insert size of L-libraries were, 6 and 7 bp shorter in the commercial cfDNA reference and real clinical cfDNA samples respectively than in their matched R-libraries (Figure 1C). The above results indicated that a large proportion of magnetic beads used in the process of library construction could increase the shorter fragment ratio of the insert DNA in both commercial cfDNA references and real clinical cfDNA samples.
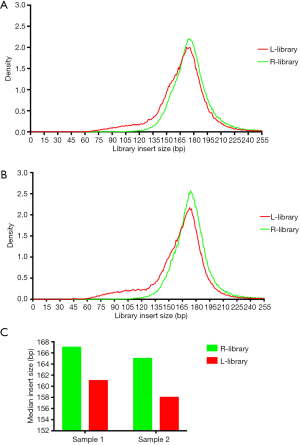
L-library could enrich shorter cfDNA and increase ctDNA detection
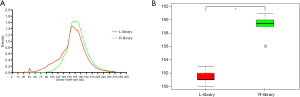
To determine whether the small fragments of ctDNA were enriched, we used plasmids which contained some known mutations (10 mutant sites) and inserted them into the fragments with lengths shorter than 150 bp using ultrasonic instruments to simulate the ctDNA (the fragment distribution is shown in Figure S2). Then, these fragmented DNA plasmids were spiked into the gDNA cell line (GM10840) which was extracted and fragmented using micrococcal nuclease and cfDNA to produce three different frequencies of gradient samples: simulation sample 1 with a plasmid dilution of 10–5, simulation sample 2 with a plasmid dilution of 5×10–7, and simulation sample 3 with plasmid dilution of 5×10–8. We speculated that the L-library might retain the shorter fragments derived from the plasmids, which meant the variant allele frequency might be higher in the L-library than in the R-library. Then, the L- and R-library were constructed to confirm if the large proportion of magnetic beads could recover more plasmid-derived short DNA and thus increase the sensitivity. A density plot was used to visualize the insert size distribution of both library types (Figure 2A), and the median insert size of the L-library was found to be significantly shorter than that of the R-library (Figure 2B).
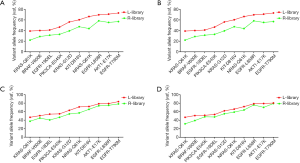
Next, we analyzed the variant allele frequency of the 10 sites for each sample. For sample 1 (10–5), when using a 20 ng or 5 ng DNA input, the 10 mutations were detected in both the L-and R-library, but the frequency in the L-library was significantly higher than that of the R-library for all 10 mutations (Figure 3A,B). Specifically, the average variant allele frequency detected by the L-library was 56.58% and ranged from 39.07% to 73.34%, while the average variant allele frequency detected by the R-library was 41.69% and ranged from 18.27% to 59.62% (P<0.05, Table S3). Apart from the for BRAF V600E mutation when 5 ng of cfDNA input was used (Figure S3), the sample 2 (5×10–7) results were the same: the average variant allele frequency detected by the L-library was 5.69% and ranged from 1.60% to 12.77%, while the average variant allele frequency detected by the R-library was 2.65% and ranged from 0.64% to 5.34% (P<0.05, Table S3). We also focused on the short fragment sizes ranging from 90 bp to 150 bp; it has been reported in a number of studies that only analyzed or enriched DNA insertion of this range could improve the ctDNA detection. We found that the frequency calculated with a 90–150 bp insert size reads was higher than that with all-size reads, especially reads >150 bp (Table S4), due to an assay design in which almost mutation fragments were shorter than 150 bp. So, we were more concerned with the impact of the L- and R-library in this range. The results indicated that the frequencies calculated with 90–150 bp insert size reads were higher in the L-library than in the R-library with both a 20 ng and 5 ng input in sample 1 (10–5) (Figure 3C,D), sample 2 (5×10–7), and sample 3 (5×10–8) (data not shown). For sample 3 (5×10–8), when 20 ng of DNA input was used, 8 and 2 of the 10 sites were detected in the L- and R-library, respectively (Figure 4A), and the frequencies of the two sites detected by the R-library had lower variant allele frequency than those detected by the L-library (0.68% and 0.62% vs. 1.27% and 0.71%, respectively). In addition, three mutations lower than 0.5% were detected in the L-library (Table S3). In particular, when the input was reduced to 5 ng, 5 and 1 of the 10 sites were detected in the L- and R-library, respectively (Figure 4B). The L-library not only increased the frequency of plasmid-derived mutation detection, but also improved the detection sensitivity for low frequencies.

Clinical samples showed that fragments 91–150 bp in size could improve detection sensitivity
To investigate if the cfDNA of cancer patients yielded the same results in the L-library, we tested 26 clinical cfDNA samples (27 mutations) which had been verified by NGS panel (30 ng of cfDNA, KAPA HyperPrep kit, NovoPM panel), and L-libraries with different cfDNA input amounts were constructed. The findings showed that the frequencies of 16 mutations were higher than those of the R-library, while 11 mutation frequencies were lower than those of the R-library (Figure 5A). Although the numbers of higher variant allele frequency in the L-library were higher than those in the R-library for the 27 mutations, the statistical analysis showed (Figure 5B) that there was no significant difference.

Since the L-library construction used a large proportion of magnetic beads, the inserted fragment size was continuous, which made it possible to analyze the contribution of different fragment length ranges to the results. Previous methods have generally directly used the Pippin system to recover the inserted fragments in the 90–150 bp range, or have used bioinformatics methods to analyze reads in this length range. Thus, all lose or partially lose the ability to analyze fragments of other lengths, especially small fragments, as these small fragments can be lost during the library construction.
Here we analyzed the length ranges of 30–90, 91–150, and >150 bp to investigate the distribution of the different lengths. We first analyzed the proportion of reads in different fragment ranges. As shown in Figure 6A, the average reads ratios of 30–90, 91–150, and >150 bp fragments were 5.35%, 29.20%, and 65.45%, respectively, which accorded with the preference of magnetic beads for binding larger fragments.
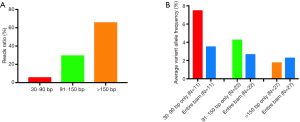
We then analyzed the reads of different fragment ranges and found that 11 of the 27 mutants could provide the alteration reads (1–10 reads, average 3.1 reads) in the 30–90 bp range. The average calculated frequency of these 11 mutations with 30–90 bp fragments was 7.45%, and when the entire BAM was used for analysis, the average frequency was 3.49% (Figure 6B). Similarly, in the range of 91–150 bp, 22 of 27 mutants could provide the alteration reads, and the average calculated frequencies of these 22 mutations with 91–150 bp fragments and the entire BAM were 4.23% and 2.64%, respectively (Figure 6B). In the analysis of the fragments >150 bp, all 27 mutants could provide the alteration reads, but there was no significant difference in the average frequency calculated with >150 bp and the entire BAM (1.74% vs. 2.26%, respectively) (Figure 6B). These results showed that the content of ctDNA was higher in the small fragment range, and when only short fragments were analyzed, the high background of non-mutant cfDNA was reduced, leading to a higher frequency. However, when the >150 bp fragments were analyzed, the cfDNA derived from healthy tissue formed a large non-mutational background; otherwise, the ctDNA was usually degraded and shorter, so we deduced that the average frequency calculated with the >150 bp fragments was lower than that with all fragments.
Using the ssDNA L-library method we could increase the opportunity to obtain alteration reads from short fragments, which was important for low variant allele frequency detection. It may provide an approach for acquiring short fragments from cfDNA and could be applied to the research of ctDNA fragment characteristics.
Discussion
In 1989, Stroun found that the increased cfDNA in the plasma of cancer patients had similar physical characteristics to tumor tissue DNA. It was not until 1996, with the development of sequencing technology, that several groups of researchers were able to simultaneously identify tumor tissue as the source of significantly increased levels of cfDNA in cancer patients. Samples can be obtained from blood or other body fluids and can be non-invasive which make ctDNA detection using in the tumor diagnosis, guiding treatment and prognosis evaluation is easier to accept. However, ctDNA detection has some limitations, its content in blood or body fluid is very low; Secondly, because of the heterogeneity of tumor cells, ctDNA technology is very demanding. So the detection of cfDNA for non-invasive diagnosis requires higher sensitivity and accuracy (8,9,22). Through the development of an array of techniques, such as large-scale parallel sequencing and ultra-deep sequencing (e.g., Duplex sequencing and Integrated digital error suppression-enhanced CAPP-seq), the sensitivity and specificity of ctDNA assays have been improved significantly (7-9).
Regular library construction uses dsDNA and a normal ratio of magnetic beads for gDNA and formalin-fixed paraffin-embedded (FFPE) samples, which are usually fragmented to 200–300 bp to fit with the Illumina PE150 sequence strategy. Therefore, traditional library construction methods are subject to losing a considerable number of short DNA templates, especially fragments shorter than 150 bp. Because the cfDNA derived from tumor cells is prone to fragmentation and degradation, normal library methods may lose ctDNA, which can increase the difficulty in detection. Although previous studies have provided evidence that enrichment of cfDNA fragments ranging from 90–150 bp can increase the ctDNA detection sensitivity (16-21), no details have been provided concerning the role of the different fragment sizes in detection. One of the main reasons for this is that in previous methods, substantial amounts of short DNA were lost, especially during the ligation and post-cleanup step when the short adapter was used. In our study, a large proportion of magnetic beads was used to perform the purification steps enabling the enrichment of small fragments and creating a more continuously graded library, beginning from very small fragments.
As mentioned in the introduction section, there are mainly three methods used to improve ctDNA detection.
The first method usually requires extra instruments and extensive time, with the size selection before library construction leading to a very low amount of cfDNA input, which could cause the high duplication (17). Alternatively, size selection after library preparation may provide several advantages over the pre-amplification size selection method. First, the intact input can produce more material after amplification, which then reduces the loss of rare mutant fragments. Second, the fragment size can be increased by ~130 bp after adapter ligation, which makes it easier to separate by conventional size-selection methods (e.g., electrophoresis and the Pippin system). Third, the libraries with different indices could perform size selection together, making the process highly scalable and reducing cost.
The second method is a bioinformatics approach, which uses 90–150 bp sized fragments for analysis. Here there were two limitations: if 90–150 bp or shorter fragments are lost during the library preparation, few reads are left available for analysis; furthermore, the 90–150 bp insert fragments may be insufficient for read depth.
The third method is ssDNA library preparation, which can enrich more ctDNA than the dsDNA method. However, the developers of this method reported that the extent of increased ctDNA content was very limited, and suggested to combine the ssDNA method with other methods such as size selection to increase the ctDNA content (21).
In the present study, we used ssDNA with a large proportion of magnetic beads in an attempt to improve the enrichment and detection of ctDNA. However, several limitations and conflicts were evident. Firstly, the samples number was small, with only 26 patient samples being used, and mostly at insufficient amounts. Secondly, not all the frequencies detected by the ssDNA L-library were higher than the prior frequencies detected by NovoPM (dsDNA). The reason for this might be that not all the short DNA fragments enriched by the L-library were mutated, which was different from the plasmid simulation. We also believe that there were a large number of unmutated fragments in the ctDNA, the amount of which varied according to the cancer type and cancer stage. Indeed, it has been largely established by different studies that the fraction of circulating DNA derived from the tumor can significantly vary according to tumor types, disease burden and tumor stage. This data suggests the potential diagnostic and prognostic values of ctDNA. Currently, different effective molecularly targeted therapies, including EGFR, ALK and BRAF inhibitors, are approved for NSCLC patients. The use of highly sensitive technique for ctDNA genotyping can allow detection of an increasing number of targetable oncogenic mutations, also when present at low frequency, and personalize treatment and can offer the possibility to monitor the emergence of resistance due to acquisition of secondary molecular alterations. Thirdly, we compared the ssDNA L-library, dsDNA, and 3D PCR data, and the variant allele frequencies were 4.97%, 2.22%, and 3.65%, (sample 17) and 4.35%, 3.93%, and 3.92% (sample 18), respectively. The frequencies of the L-library were higher than those of 3D PCR, possibly because 3D PCR requires a certain length of amplified fragments, and shorter fragments with no matched primers could not be detected. Finally, the amount of input in the ssDNA L-library was lower than that of the dsDNA test, which might have caused some discrepancies in the results. A similar phenomenon occurred in the plasmid simulation test: the frequency of the BRAF V600E mutation was lower in the L-library than in the R-library.
Acknowledgments
The authors appreciate the academic support from the AME Cancer Biology Collaborative Group.
Funding: This work was supported by Henan Province Health and Youth Subject Leader Training Project {[2020]60}; Leading Talent Cultivation Project of Henan Health Science and Technology Innovation Talents (YXKC2020009); ZHONGYUAN QIANREN JIHUA; Henan International Joint Laboratory of Drug Resistance and Reversal of Targeted Therapy for Lung Cancer {[2021]10}; Henan Medical Key Laboratory of Refractory Lung Cancer {[2020]27}; Henan Refractory Lung Cancer Drug Treatment Engineering Technology Research Center {[2020]4}; the 51282 Project Leading Talent of Henan Provincial Health Science and Technology Innovation Talents {[2016]32}; Huilan Charity Funda Project (HL-HS2020-129).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-21-180
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-21-180
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-21-180). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was conducted in accordance with the ethical guidelines of the United States’ common rule, and the protocol was approved by the Research Ethics Committee of Henan Cancer Hospital. All the patients in the present study have signed informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cheng F, Su L, Qian C. Circulating tumor DNA: a promising biomarker in the liquid biopsy of cancer. Oncotarget 2016;7:48832-41. [Crossref] [PubMed]
- Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 2017;14:531-48. [Crossref] [PubMed]
- Fernandes Marques J, Pereira Reis J, Fernandes G, et al. Circulating tumor DNA: a step into the future of cancer management. Acta Cytol 2019;63:456-65. [Crossref] [PubMed]
- Wan JCM, Massie C, Garcia-Corbacho J, et al. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 2017;17:223-38. [Crossref] [PubMed]
- Lu L, Bi J, Bao L. Genetic profiling of cancer with circulating tumor DNA analysis. J Genet Genomics 2018;45:79-85. [Crossref] [PubMed]
- Volckmar AL, Sültmann H, Riediger A, et al. A field guide for cancer diagnostics using cell-free DNA: From principles to practice and clinical applications. Genes Chromosomes Cancer 2018;57:123-39. [Crossref] [PubMed]
- Bai Y, Wang Z, Liu Z, et al. Technical progress in circulating tumor DNA analysis using next generation sequencing. Mol Cell Probes 2020;49:101480 [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547-55. [Crossref] [PubMed]
- Desai A, Kallianpur S, Mani A, et al. Quantification of circulating plasma cell free DNA fragments in patients with oral cancer and precancer. Gulf J Oncolog 2018;1:11-7. [PubMed]
- Giacona MB, Ruben GC, Iczkowski KA, et al. Cell-free DNA in human blood plasma: length measurements in patients with pancreatic cancer and healthy controls. Pancreas 1998;17:89-97. [Crossref] [PubMed]
- Jiang P, Chan CW, Chan KC, et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc Natl Acad Sci U S A 2015;112:E1317-25. [Crossref] [PubMed]
- Mouliere F, El Messaoudi S, Pang D, et al. Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Mol Oncol 2014;8:927-41. [Crossref] [PubMed]
- Mouliere F, Robert B, Arnau Peyrotte E, et al. High fragmentation characterizes tumour-derived circulating DNA. PLoS One 2011;6:e23418 [Crossref] [PubMed]
- Umetani N, Giuliano AE, Hiramatsu SH, et al. Prediction of breast tumor progression by integrity of free circulating DNA in serum. J Clin Oncol 2006;24:4270-6. [Crossref] [PubMed]
- Underhill HR, Kitzman JO, Hellwig S, et al. Fragment length of circulating tumor DNA. PLoS Genet 2016;12:e1006162 [Crossref] [PubMed]
- Mouliere F, Chandrananda D, Piskorz AM, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med 2018;10:eaat4921 [Crossref] [PubMed]
- Cristiano S, Leal A, Phallen J, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019;570:385-9. [Crossref] [PubMed]
- Hellwig S, Nix DA, Gligorich KM, et al. Automated size selection for short cell-free DNA fragments enriches for circulating tumor DNA and improves error correction during next generation sequencing. PLoS One 2018;13:e0197333 [Crossref] [PubMed]
- Liu X, Liu L, Ji Y, et al. Enrichment of short mutant cell-free DNA fragments enhanced detection of pancreatic cancer. EBioMedicine 2019;41:345-56. [Crossref] [PubMed]
- Zhu J, Huang J, Zhang P, et al. Advantages of Single-Stranded DNA Over Double-Stranded DNA Library Preparation for Capturing Cell-Free Tumor DNA in Plasma. Mol Diagn Ther 2020;24:95-101. [Crossref] [PubMed]
- Reinert T, Schøler LV, Thomsen R, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016;65:625-34. [Crossref] [PubMed]
(English Language Editor: J. Gray)


