
circFBXW7 attenuates malignant progression in lung adenocarcinoma by sponging miR-942-5p
Introduction
Lung cancer still ranks among the most common malignancies worldwide, and remains the primary cause of cancer-related death in men and women (1). Small-cell lung cancer (SCLC) and non-small-cell lung cancer (NSCLC) constitute the two primary types of lung cancer, with approximately 85% of lung cancer cases being NSCLC (2). The two main NSCLC histological subtypes are lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (3). Despite continuous advancement in surgical therapy, radiochemotherapy and targeted therapy for lung cancer over the past 20 years, the 5-year survival rate in lung cancer patients is only 20–30% (4). As LUAD has become the most common subtype in NSCLC (5), new methods that can prolong the survival time for the LUAD patients are needed. Gene aberration plays an important role in occurrence and development of tumors. For instance, programmed death-ligand 1 (PD-L1) can predict the prognosis of the LUAD (6), while epidermal growth factor receptor (EGFR) and anaplastic lymphoma kinase (ALK) have been used for targeted gene therapy in LUAD treatment (6). Most of the recent research related to gene aberration has focused on noncoding RNA (ncRNA) (7,8), particularly circular RNA (circRNA) (9).
As a specific kind of long noncoding RNA, circRNA is generated by back-splicing with either a 5’-3’ polarity or a polyadenylous tail and is characterized by a covalently closed loop (10). circRNAs function as key genes in the tumorigenesis of cancers, influencing proliferation, metastasis, apoptosis, autophagy, and other processes. circRNAs can resist multiple exonuclease and are much more stable than linear RNAs. Furthermore, circRNAs are related to a specific cell type, diseases, or development stage in different conditions, indicating their potential as biomarkers for the diagnosis and therapy of diseases. As a tumor suppressor, circFBXW7 is produced by the transcription and splicing of F-box and WD repeat domain containing 7 (FBXW7). circFBXW7 suppresses cell proliferation and metastasis in glioma (11) and may be utilized as a potential therapeutic target (12). Ye et al. (13) found that circFBXW7 could also inhibit malignant progression in triple-negative breast cancer.
The proliferation and metastasis represent significant processes in cancer progression; however, whether circFBXW7 function in LUAD tumor progression influences proliferation or metastasis has not been established. Simultaneously, many specific pathways involved in this procedure, especially epithelial-mesenchymal transition (EMT) pathway. In both physiological and pathological processes, EMT exerts its effects in various activities, including wound healing, tumor metastasis (14), and embryonic organ development (15). In tumor metastasis (16), as opposed to having normal characteristics, such as intercellular adhesion and cell polarity, the epithelial cells are characterized by altered shape, increased motility, invasive properties, and migratory potential, all of which represent a mesenchymal phenotype caused by EMT (17).
In the present study we analyzed whether circFBXW7 may influence proliferation and metastasis of lung cancer cells and impact patient survival. We performed in vitro and in vivo experiments to evaluate the impact of circFBXW7 in LUAD development, explored by bioinformatic analysis the involved microRNAs and genes and performed functional validation of the in silico results. In summary, we verified that circFBXW7 impacts patient prognosis and attenuates malignant progression in LUAD through regulation of the miR-942-5p/BARX2 axis, which is involved in the EMT pathway, providing a potential target therapy for LUAD. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/tlcr-21-230).
Methods
Cell lines and cell culture
Human lung normal epithelial cell lines, BEAS-2B, and LUAD cell lines, A549, HCC2279, and H1299 acquired from the Cell Bank of Chinese Academy of Sciences were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, USA) mixed with 100 U/mL penicillin, 100 µg/mL streptomycin, L-glutamine, and 10% fetal bovine serum (FBS, ExCell Bio, China) at 37 °C in a humidified incubator with 5% CO2.
Tissue samples
After receiving informed consent for use of tissue samples, 40 intraoperative fresh LUAD tissue specimens and paired adjacent normal lung tissues were collected from patients in the Affiliated Hospital of Qingdao University between January 2015 and December 2015. The median follow up was 63.5 months. The diagnosis of histology and pathology for LUAD patients was based on the 2015 World Health Organization (WHO) classification. Tissue samples from patients receiving adjuvant therapy and neoadjuvant therapy were excluded. The main clinical characteristics of the patients are summarized in Table 1. The samples were infiltrated by RNAwait (Solarbio, China) at 4 °C overnight and stored at –80 °C before RNA extraction and quantitative real-time reverse transcriptase PCR (qRT-PCR). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of the Affiliated Hospital of Qingdao University (No. QYFYKY 2018-10-11-2).
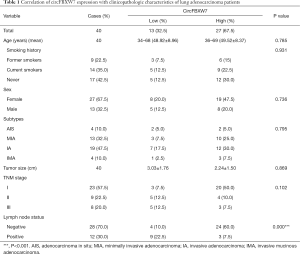
Full table
RNA extraction and qRT-PCR
RNA was acquired by Trizol (Thermo Fisher Scientific, USA). Complement DNA (cDNA) synthesis and qRT-PCR analysis were performed by PrimeScript™ RT reagent Kit (Perfect Real Time) and TB Green Fast qPCR Mix (Takara, China). GAPDH and U6 were used as the internal reference to examine the expression level of circFBXW7, FBXW7 mRNA, BARX2 mRNA, miR-668-3p, miR-887-3p, and miR-942-5p. The Primers involved are listed under Additional Files in Table S1. The sequence of circFBXW7 was shown in Table S2. The expression of each gene was calculated by the 2−ΔΔCt method. qRT-PCR analysis was repeated 3 times.
Nuclear and cytoplasmic RNA isolation assay
The PARIS Kit (Invitrogen, USA) was used to perform cytoplasmic and nuclear RNA isolation. After being lysed in cell fractionation buffer, A549 cell lysates were centrifuged, and the supernatant was gathered. Cell fractionation buffer was used to rinse the rest of the lysates, and the lysates were centrifuged. Cell disruption buffer was used to lyse the nuclei. The expression of genes was assessed through qRT-PCR based on isolated RNAs. The experiment was repeated 3 times.
Fluorescence in situ hybridization
Cells were cultured on coverslips overnight, fixed in 4% paraformaldehyde and permeablized in 0.1% Triton X-100 for 15 minutes at room temperature. Subsequently, cells were treated with 2× SSC (saline sodium citrate) for 30 minutes at 37 °C in an incubator. Then, the cells were hybridized in hybridization buffer (18) with Cy3-labeled circFBXW7 probes (5'-Cy3-TATCCTAAGGAAGTAATCTTTTGTTGTTTTTGTAT-3') at 37 °C overnight. 4,6-diamidino-2-phenylindole (DAPI) was used to counterstain the nuclei of cells. Then the figures were captured on a laser confocal scanning microscope (Leica, Germany). Experiments were repeated 3 times.
Transiently and stably overexpressing circFBXW7
To acquire A549 and HCC2279 cell lines transiently overexpressing circFBXW7, we first cloned the full length of 1,127-bp circFBXW7 into a plasmid cloning DNA (pcDNA) 3.1(+) expression vector (HanBio, China) between the restriction enzyme EcoR I (GAATTC) to generate a circFBXW7 overexpression vector construct. A549 and HCC2279 cells was transiently transfected with the constructed vector using Lipofectamine 3000 (Thermo Fisher Scientific). The empty vector pcDNA 3.1(+) served as negative control. After transfection for 24 or 48 hours, cells were collected for additional experiments. miR-942-5p mimics and miR-942-5p NC were designed and synthesized by GeneChem Co (China).
To establish A549 cells stably overexpressing circFBXW7, we infected A549 cells with human B-lymphotropic virus-circFBXW7 (HBLV-circFBXW7) or human B-lymphotropic virus-negative control (HBLV-NC) lentiviruses (HANBIO, China). After being cultured for 3 days, A549 cells were exposed to 0.5 µg/mL of puromycin (HANBIO).
RNA interference
circFBXW7 was specifically knocked down using small interfering RNAs (siRNAs) (si-circFBXW7-1, 5'-CAACAAAAGAUUACUUCCU-3'; si-circFBXW7-2, 5'-CAAAAGAUUACUUCCUUAG-3') (GeneChem), which were taken from the CircInteractome database (http://circinteractome.nia.nih.gov) and synthesized by GenePharma (China). A549 and HCC2279 cells were transfected with 100 pmol of siRNA using Lipofectamine 3000 (Thermo Fisher Scientific). siRNA-NC (5'-AGACCUAUACAUCUAUAUG-3') was used as a negative control (NC). Cells were collected 24 hours after transfection for qRT-PCR of circFBXW7, FBXW7 mRNA, miR-942-5p, miR-778-3p, miR-668-3p, and BARX2 mRNA expression or for other experiments. si-BARX2 (5'-AUACCAGGUCUUCACCUGC-3') was also synthesized by GenePharma.
MTT assay
To evaluate proliferative ability, we seeded A549 and HCC2279 cells in 96-well plates and examined them via MTT assay (Sigma-Aldrich, USA). Cells were then cultured in 500 µg/mL MTT solution for 4 hours. We then removed the MTT solution and added 100 µL of dimethyl sulfoxide (DMSO) per well into the 96-well plates. After being vibrated for 60 seconds, the absorbance of 450 nm was monitored via a fully functional microporous plate detector (BioTek, USA). Each experiment was carried out 3 times.
EdU assay
A549 and HCC2279 cell lines were seeded in 96-well plates overnight. EdU kit (RiboBio, China) was utilized for assessing cell proliferative ability. Images were captured by an inverted fluorescence microscope (Leica). Nuclei were redyed with Hoechst staining (RiboBio) to assess cell viability. The results were analyzed by ImageJ software (National Institutes of Health, USA). The cell viability was measured as the percentage of cells that were stained with Hoechst. Each experiment was conducted 3 times.
Wound healing assay
A549 and HCC2279 cells were seeded onto six-well plates and cultured overnight. A linear wound was created within the confluent monolayers by scratching the cells with sterile 1-mL pipette tips (time 0 h). Then the wound was washed by phosphate buffer saline twice. Cells were further cultured with medium containing 5% FBS in 48 h. Cell migration was imaged at 0 and 48 h after injury, by using 10 high-power fields with an inverted microscope. Remodeling was measured as diminishing distance across the induced injury, standardized to the 0 h control, and recorded as relative migration. Each experiment was conducted 3 times.
Cell migration assay
The migration ability of cells was evaluated via Transwell chamber (8 µm Costar; Corning, USA). After being resuspended in serum-free medium, cells were seeded onto the upper chamber with a density of 2×105 cells per chamber overnight. Then, the chamber was fixed with 4% paraformaldehyde for 40 minutes and stained with 5% crystal violet stain for 30 minutes. Treated cells were observed and counted using an inverted fluorescence microscope (Leica). We chose 5 random views and calculated the average number of cells in each chamber. The results were analyzed by ImageJ software. Each experiment was conducted 3 times.
Prediction of miRNA targets
Target genes of circRNA were predicted with CircInteractome (https://circinteractome.irp.nia.nih.gov/), CircBank (http://www.circbank.cn/) and StarBase (http://starbase.sysu.edu.cn/) databases.
RNA pull-down assay
In brief, we first collected the cell lysates treated with cell lysis. The sequence of the circFBXW7 probe was the following: 5'-TATCCTAAGGAAGTAATCTTTTGTTGTTTTTGTAT-Biotin-3'. Streptavidin-coated agarose beads were incubated with the biotin-labeled circFBXW7 probe at 4 °C for 2 hours to acquire probe-coated beads. Lysates of A549 and HCC2279 cells were incubated with probe-coated beads for 3 hours at 4 °C. Then, the RNA complexes binding to the beads were collected. The expression levels of miR-942-5p, miR-778-3p, miR-668-3p, and circFBXW7 were analyzed by qRT-PCR. U6 and GAPDH were selected as negative controls. Each experiment was carried out 3 times.
Dual-luciferase reporter assay
The potential binding site of circFBXW7 or BARX2 and miR-942-5p was replaced by circFBXW7-mutant (Mut) and BARX2-3'UTR-Mut. A549 and HCC2279 cells were seeded in a 96-well plate overnight. When the cells obtained 50–70% confluence, they were co-transfected with circFBXW7-Wt (or circFBXW7-Mut) or BARX2-3'UTR-Wt (or BARX2-3'UTR-Mut), and miR-942-5p mimics or miR-942-5p NC. The luciferase activities were tested by a fully functional microporous plate detector (BioTek) 48 hours after transfection. Each experiment was performed 3 times.
RNA immunoprecipitation assay
miR-942-5p mimics and miR-942-5p NC were transfected into A549 and HCC2279 cells. After being lysed, the cell lysate was mixed with treated beads and rotated for 4 hours at 4 °C. Then, the AGO2-specific antibody and immunoglobin G (IgG) antibody were used in AGO2 immunoprecipitation. Beads were subsequently washed 6 times in immunoprecipitation buffer (150 mM KCl, 25 mM Tris-HCl, pH 7.4, 5 mM EDTA, 0.5% Triton X-100, and 5 mM DTT supplemented with RNase inhibitor and proteinase inhibitor cocktail), and the RNA was collected using Trizol (Invitrogen) to perform qRT-PCR. Each experiment was conducted 3 times.
Western blot analysis
Protein was purified and used for western blot analysis as previously described (19). The antibodies involved were as follows: mouse anti-BARX2 (Abcam, UK); rabbit anti-E-cadherin, anti-N-cadherin, and anti-Vimentin (ABclonal, China); rabbit anti-β-actin, and anti-rabbit and anti-mouse secondary antibodies (ABclonal). The molecular sizes shown on the immunoblots of BARX2, E-cadherin, N-cadherin, Vimentin, and β-actin were 52, 125, 140, 57, and 43 kD, respectively. Each experiment was performed 3 times.
Tumor xenograft models
After gaining approval for animal experiments from the Affiliated Hospital of Qingdao University, we used 12 female BALB/c nude mice (5–6 weeks of age, 18±2 g; Pengyue Laboratory Animal Technology, China) as xenograft models. Nude mice were randomly and equally divided into two groups: a HBLV-NC group and a HBLV-circFBXW7 group. They were bred in a specific pathogen-free laboratory, with 6 mice per transparent cage, and in a stable light-dark cycle. Then, 5×106 cells were subcutaneously injected into the left flanks of the mice in the laboratory in the morning of the third day after their move. The tumor volume was calculated as 0.52× (length × width2) and recorded every week; 35 days after injection, the mice were euthanatized. The tumor volume and tumor weight were recorded and analyzed by t-test. The tumors were then subjected to immunohistochemistry staining. Experiments were performed under a project license (No. 1107262011003592) granted by ethics board of the Affiliated Hospital of Qingdao University, in compliance with national guidelines for the care and use of animals.
Statistical analysis
Results are presented as mean ± standard deviation (SD) and percentage. Paired or unpaired t-test (two-tailed) and χ2 analysis were used to analyze the differences between the two groups. The association between two groups was assessed by Pearson’s correlation test. X-tile was used to measure the cutoff value of the expression level of the circFBXW7 in patients, and the cutoff value was 0.8. A P value <0.05 was considered significant. GraphPad Prism 8.0 software (GraphPad, USA) was used to perform the statistical analyses. Overall survival (OS) was defined as the time from surgery until death from the disease. Disease free survival (DFS) was defined as the time from surgery until recurrence from the disease. Both DFS and OS were analyzed by the Kaplan-Meier analysis and log-rank tests using R software (R Foundation for Statistical Computing, Austria), based on “survival” and “survminer” packages.
Results
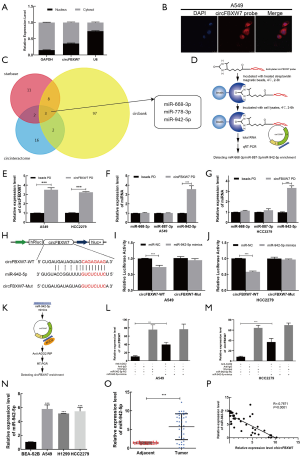
circFBXW7 was downregulated in LUAD and correlated with worse clinical outcomes
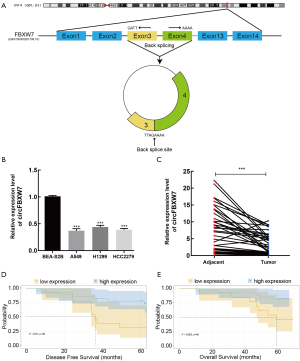
According to the CircBase and CircBank databases, circFBXW7 (CircBase ID: has_circ_0001451) is derived from exons 3 and 4, with their intermediate intron of FBXW7 located on chromosome 4q31.21 (Figure 1A). Compared to the normal lung cell lines (Figure 1B) and adjacent tumor tissues (Figure 1C), the expression level of circFBXW7 was lower than the expression level in LUAD lines and LUAD tissues, which was tested by qRT-PCR. After examining its expression level, we then chose A549 and HCC2279 cell lines for the subsequent experiments owing to their lower expression level. To verify the association between the circFBXW7 expression level and clinical characteristics, we analyzed the circFBXW7 expression in 40 LUAD tumor samples. For the prognosis analysis patients were divided into two groups based on the cutoff obtained with X-tile: a high expression level group and a low expression level group. Kaplan-Meier survival analysis and log-rank test revealed that LUAD patients with low expression levels of circFBXW7 had shorter DFS (Figure 1D) and OS (Figure 1E). Median DFS for patients with high levels was 63 months (95% CI: 45.69–60.76), while it was 36 months (95% CI: 30.30–54.09) for those with low levels. Median OS for patients with high levels was 65 months (95% CI: 54.09–64.65), while it was 59 months (95% CI: 43.08–63.53) for those with low levels. Furthermore, the clinical characteristics indicated that the low expression level of circFBXW7 was correlated with positive lymph node metastasis in LUAD patients (Table 1).
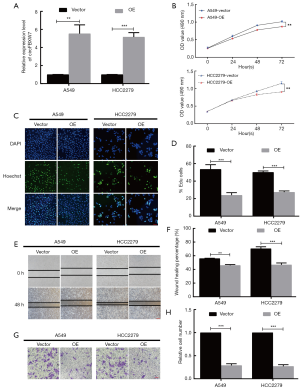
Overexpression of circFBXW7 attenuated the proliferation and metastasis of LUAD cells
We verified the transfection efficiency of the overexpression vector of circFBXW7 in A549 and HCC2279 cell lines (Figure 2A). MTT assays (Figure 2B) and EdU assays (Figure 2C,D) showed that the proliferation ability in A549 and HCC2279 cell lines was significantly decreased at 72h by the upregulation of circFBXW7. To further investigate the impact of the overexpression vector of circFBXW7 on the migration capacity of A549 and HCC2279 cells, we performed wound-healing assays (Figure 2E,F) and Transwell assays (Figure 2G,H), which suggested that the cells migrated less than the normal cells after overexpressed the circFBXW7. Our results demonstrated that the upregulation of circFBXW7 significantly attenuated the migration ability of A549 and HCC2279 cell lines.
Knockdown of circFBXW7 facilitated the proliferation and migration of LUAD cells
In addition to knocking down circFBXW7 in LUAD cell lines, we applied si-circFBXW7-1/2 and verified its transfection efficiency in A549 and HCC2279 cell lines and excluded the influence to FBXW7 mRNA of si-circFBXW7-1/2 (Figure 3A). MTT assays (Figure 3B,C) and EdU assays (Figure 3D,E) consistently showed that the downregulation of circFBXW7 augmented cell proliferation of A549 and HCC2279 cell lines. Consistent with the above results, after knockdown of circFBXW7, the A549 and HCC2279 cell lines showed stronger migration ability in wound-healing assays (Figure 3F,G) and Transwell assays (Figure 3H,I).

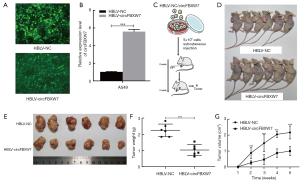
The overexpression of circFBXW7 attenuated tumor proliferation in vivo
In order to clarify the biological function of circFBXW7 in vivo, we acquired the lentivirus stably overexpressing circFBXW7, which contained HBLV-circFBXW7 plasmids, while the negative control group contained HBLV-NC. After infection with the lentivirus, we acquired the A549 cells stably overexpressing circFBXW7 (HBLV-circFBXW7) and negative control A549 cells (HBLV-NC) (Figure 4A). The expression of circFBXW7 in HBLV-circFBXW7 cells was 5 times greater than that in HBLV-NC cells (Figure 4B). Then, BALB/c nude mice were injected with HBLV-circFBXW7 cells and HBLV-NC cells (Figure 4C). The original pictures of Figure 4C were provided by Servier Medical Art (https://smart.servier.com) under CC BY 3.0 license. No adverse reactions occurred in mice. Five weeks after being inoculated, the mice were euthanized and surgically dissected (Figure 4D,E). As expected, the mice inoculated with HBLV-circFBXW7 cells exhibited a significant reduction in tumor weight and volume (Figure 4F,G).
circFBXW7 acted as a sponge of miR-942-5p
After nuclear and cytoplasmic RNA was isolated from cells, qRT-PCR analysis revealed that circFBXW7 was mostly localized in the cytoplasm of A549 cells (Figure 5A). Representative images of endogenous circFBXW7 in A549 cells obtained by fluorescence in situ hybridization suggested that circFBXW7 was mainly located in the cytoplasm of A549 cells (Figure 5B). We thus hypothesized that circFBXW7 may serve as a miRNA sponge. According to the Circular RNA Interactome website (https://circinteractome.nia.nih.gov/index.html), StarBase database (http://starbase.sysu.edu.cn/), and CircBank database (http://www.circbank.cn/searchCirc.html), miR-668-3p, miR-887-3p, and miR-942-5p were screened as the potential target binding genes of circFBXW7 in the intersection of the 3 databases (Figure 5C). RNA pull-down analysis (Figure 5D,E) showed the higher expression level of miR-942-5p in the RNA complexes and suggested that endogenous miR-942-5p was connected with biocatalytic probes against circFBXW7 in A549 (Figure 5F) and HCC2279 (Figure 5G) cell lines, confirming the relationship between circFBXW7 and miR-942-5p. We then applied dual luciferase reporter assay by inserting the wild-type or miR-942-5p binding site mutant of circFBXW7 (Figure 5H). The relative luciferase activity of circFBXW7 wild type (circFBXW7-WT) reporter was reduced more than 30% in the cells transfected with miR-942-5p mimics compared to negative control group in the A549 (Figure 5I) and HCC2279 cell lines (Figure 5J), whereas this reduction was not observed in the miR-942-5p mutated binding site. Finally, as miRNAs always act in an AGO2-dependent manner, we performed an RNA immunoprecipitation (RIP) assay (20) (Figure 5K). The results indicated that circFBXW7 was significantly enriched in the miR-942-5p mimics group, which indicated that circFBXW7 directly interacted with miR-942-5p in A549 (Figure 5L) and HCC2279 (Figure 5M) cell lines. In addition, we found the upregulation of miR-942-5p in LUAD cell lines and cancer tissues (Figure 5N,O). Pearson correlation analysis indicated that the expression level of miRNA-942-5p and that of circFBXW7 were negatively correlated to each other (R=−0.7671, P<0.0001) in tumor samples (Figure 5P).
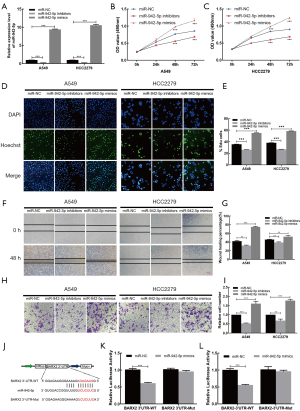
miR-942-5p decreased the expression of BARX2
We evaluated the transfection efficiency of miR-942-5p inhibitors and mimics in A549 and HCC2279 cells (Figure 6A). To investigate the proliferative activity of miR-942-5p, we performed MTT assays in A549 (Figure 6B) and HCC2279 (Figure 6C) cell lines, which indicated that miR-942-5p promoted the growth of LUAD cell lines. The results of EdU assays (Figure 6D,E) in A549 and HCC2279 cell lines were in accordance with MTT assays. Concurrently, results from wound-healing assays (Figure 6F,G) and Transwell assays (Figure 6H,I) showed that miR-942-5p could facilitate the migration ability of A549 and HCC2279 cell lines. Based on the research (21), we tried to examine the relationship of the miR-942-5p and BARX2. Additionally, dual luciferase reporter assays (Figure 6J) in A549 (Figure 6K) and HCC2279 (Figure 6L) cell lines suggested that the relative luciferase activity was predominantly inhibited after cotransfection with miR-942-5p mimics and the BARX2-3'UTR-WT vector compared with that of cells cotransfected with the BARX2-3'UTR-Mut vector.
circFBXW7 and BARX2 acted as competing endogenous RNAs in LUAD by regulating miR-942-5p
The overexpression of circFBXW7 increased the expression levels of BARX2, while this effect was reversed by the cotransfection with miR-942-5p mimics or siBARX2 in A549 and HCC2279 cell lines (Figure 7A,B). These results demonstrated that circFBXW7 and BARX2 acted as competing endogenous RNAs (ceRNAs) in LUAD by regulating miR-942-5p. MTT (Figure S1A,B) and EdU assays (Figure S1C,D,E) demonstrated that miR-942-5p mimics or siBARX2 could increase cell proliferation compared with the circFBXW7 overexpression vector alone. To further examine the influence of miR-942-5p mimics or siBARX2 on the metastatic capacity of LUAD cells, wound-healing assays (Figure S1F,G,H) and Transwell assays (Figure S1I,J,K) were also conducted. Since it has been previously shown (21) that miR-942-5p/BARX2 axis regulates EMT in NSCLC, we then performed western blot to verify the lower N-cadherin, higher E-cadherin, and lower Vimentin expression levels in A549 (Figure 7C,D) and HCC2279 (Figure 7E,F) cell lines, which suggested that the circFBXW7 inhibited the EMT pathway via miR-942-5p and BARX2.
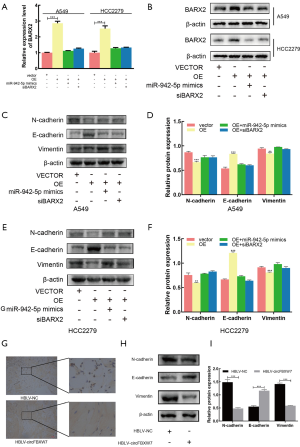
Furthermore, we assessed the higher BARX2 expression level of the tumors in the HBLV-circFBXW7 group using immunohistochemistry (Figure 7G), and assessed N-cadherin, E-cadherin, and Vimentin expression levels using western blot; these tests showed that the N-cadherin, and Vimentin were less expressed in the in vivo samples infected with the HBLV-circFBXW7. However, the expression level of E-cadherin was overexpressed in these samples (Figure 7H,I).
Discussion
In this study, we confirmed that circFBXW7 regulates proliferation and migration in NSCLC by regulation of EMT related protein through miR-942-5p/BARX2 axis. Consequently, we examined whether circFBXW7 can serve as a prognostic marker in LUAD. To our knowledge, this is the first study to explore the biological function of circFBXW7 in LUAD.
We demonstrated that the expression level of circFBXW7 in LUAD cell lines or tissues was considerably lower than that in normal cells or tissues, indicating its potential tumor-suppressing effect. In vitro biological function experiments suggested that the circFBXW7 attenuated the proliferation and migration in A549 and HCC2279 cell lines, which was in accordance with the results found in a study of glioma (11). Currently, circRNAs are recognized not as the byproduct of mRNA transcription (14), but as active molecules influencing the biological or pathological activities of the human body (22). The accumulation of circRNAs has been proven to influence carcinogenesis and cancer progression, the discovery of which has shifted our perspectives on cancer (23,24). We further proved that circFBXW7 can reduce the EMT markers, N-cadherin and Vimentin. The overexpression of circFBXW7 and the effects of circFBXW7 knockdown both indicate that circFBXW7 suppresses the proliferation and migration of LUAD.
As reported previously, circRNAs usually function as ceRNAs in tumorigenesis (25,26). However, only specific miRNAs may be bound to the appropriate binding sites with the given fragment of circRNAs (27). Therefore, we predicted the potential target miRNAs of circFBXW7 through bioinformatics analysis. Our findings from pull-down assays, dual luciferase reporter assays, and RIP assays, demonstrated a sponging interaction between circFBXW7 and miR-942-5p. The oncogenic function of miR-942-5p has been examined in many cancers (28,29). One study discovered that miR-942-5p facilitates the growth and migration of LUAD by targeting BARX2 (21). In our study, we verified the biological function of miR-942-5p in A549 and HCC2279 cell lines, which is in line with the results of the above-mentioned research. In our study, we demonstrated the relationship between miR-942-5p and BARX2. Whether transfected with miR-942-5p mimics or siBARX2, the influence of overexpressing circFBXW7 was reversed. Furthermore, the effect of circFBXW7 in EMT pathway was also decreased. These results suggest that circFBXW7 mainly exerts its negative effect on LUAD via sponging miR-942-5p and inducing BARX2 overexpression. BARX2 is related to EMT pathway (21), and EMT pathway plays an important part in cancer tumorigenesis and progression (16,30). Interference in any process of the EMT course may affect cancer progression. Clarification of the expression level of N-cadherin, E-cadherin, and Vimentin by western blot indicated that that the overexpression of circFBXW7 increased EMT in the subcutaneous tumor xenograft model. Abnormal expression of BARX2 has been reported in many cancers, including breast cancer (31) and ovarian cancer (32). In our study, we demonstrated that the expression level of BARX2 in A549 cells overexpressing circFBXW7 was higher than that in the NC group. Finally, the expression level of BARX2 in resected tumors from mice by immunohistochemical staining also indicated that BARX2 participates in the tumorigenesis and tumor progression of LUAD.
Taken together, our findings support further investigation into the relationship between circFBXW7 and other diseases. We have identified a novel potential prognostic biomarker for LUAD with potential use as a therapeutic marker. However, further basic research is needed before clinical application can be considered.
Conclusions
Our results suggest that circFBXW7 sponges miR-942-5p to suppress LUAD progression through upregulating BARX2 expression, which represents a novel mechanism in cancer progression. circFBXW7 may thus be a potential therapeutic target and prognostic biomarker for LUAD.
Acknowledgments
The authors appreciate the academic support from AME Lung Cancer Collaborative Group.
Funding: This study was funded by the Key Research and Development Programs of Shandong Province (No. 2018GSF118119) and the Qingdao Minsheng Keji Jihua, Qingdao Science and Technology Bureau (No. 17-3-3-5-nsh).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-21-230
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-21-230
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-21-230). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of the Affiliated Hospital of Qingdao University (No. QYFYKY 2018-10-11-2). Informed consent was obtained from patients. Experiments were performed under a project license (No. 1107262011003592) granted by ethics board of the Affiliated Hospital of Qingdao University, in compliance with national guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA: A Cancer Journal for Clinicians 2020;70:7-30. [Crossref] [PubMed]
- Chen Z, Fillmore CM, Hammerman PS, et al. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 2014;14:535-46. [Crossref] [PubMed]
- Langer CJ, Besse B, Gualberto A, et al. The evolving role of histology in the management of advanced non-small-cell lung cancer. J Clin Oncol 2010;28:5311-20. [Crossref] [PubMed]
- Allemani C, Matsuda T, Di Carlo V, et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 2018;391:1023-75. [Crossref] [PubMed]
- Rosell R, Karachaliou N, Arrieta O. Novel molecular targets for the treatment of lung cancer. Curr Opin Oncol 2020;32:37-43. [Crossref] [PubMed]
- Ozaki Y, Muto S, Takagi H, et al. Tumor mutation burden and immunological, genomic, and clinicopathological factors as biomarkers for checkpoint inhibitor treatment of patients with non-small-cell lung cancer. Cancer Immunol Immunother 2020;69:127-34. [Crossref] [PubMed]
- Zhang C, Ding R, Sun Y, et al. Circular RNA in tumor metastasis. Mol Ther Nucleic Acids 2021;23:1243-57. [Crossref] [PubMed]
- Zhang Y, Liu Q, Liao Q. Long noncoding RNA: a dazzling dancer in tumor immune microenvironment. J Exp Clin Cancer Res 2020;39:231. [Crossref] [PubMed]
- Liang ZZ, Guo C, Zou MM, et al. circRNA-miRNA-mRNA regulatory network in human lung cancer: an update. Cancer Cell Int 2020;20:173. [Crossref] [PubMed]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. [Crossref] [PubMed]
- Yang Y, Gao X, Zhang M, et al. Novel Role of FBXW7 Circular RNA in Repressing Glioma Tumorigenesis. J Natl Cancer Inst 2018;110:304-15. [Crossref] [PubMed]
- Li Z, Ruan Y, Zhang H, et al. Tumor-suppressive circular RNAs: Mechanisms underlying their suppression of tumor occurrence and use as therapeutic targets. Cancer Sci 2019;110:3630-8. [Crossref] [PubMed]
- Ye F, Gao G, Zou Y, et al. circFBXW7 Inhibits Malignant Progression by Sponging miR-197-3p and Encoding a 185-aa Protein in Triple-Negative Breast Cancer. Mol Ther Nucleic Acids 2019;18:88-98. [Crossref] [PubMed]
- Nishiyama M, Tsunedomi R, Yoshimura K, et al. Metastatic ability and the epithelial-mesenchymal transition in induced cancer stem-like hepatoma cells. Cancer Sci 2018;109:1101-9. [Crossref] [PubMed]
- Matoba R, Morizane Y, Shiode Y, et al. Suppressive effect of AMP-activated protein kinase on the epithelial-mesenchymal transition in retinal pigment epithelial cells. PLoS One 2017;12:e0181481 [Crossref] [PubMed]
- De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 2013;13:97-110. [Crossref] [PubMed]
- Guo M, Ehrlicher AJ, Mahammad S, et al. The role of vimentin intermediate filaments in cortical and cytoplasmic mechanics. Biophys J 2013;105:1562-8. [Crossref] [PubMed]
- Lu T, Qiu T, Han B, et al. Circular RNA circCSNK1G3 induces HOXA10 signaling and promotes the growth and metastasis of lung adenocarcinoma cells through hsa-miR-143-3p sponging. Cell Oncol (Dordr) 2021;44:297-310. [Crossref] [PubMed]
- Lu T, Qiu T, Han B, et al. Circular RNA circCSNK1G3 induces HOXA10 signaling and promotes the growth and metastasis of lung adenocarcinoma cells through hsa-miR-143-3p sponging. Cell Oncol (Dordr) 2020.
- Gregory RI, Chendrimada TP, Cooch N, et al. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 2005;123:631-40. [Crossref] [PubMed]
- Yang F, Shao C, Wei K, et al. miR-942 promotes tumor migration, invasion, and angiogenesis by regulating EMT via BARX2 in non-small-cell lung cancer. J Cell Physiol 2019;234:23596-607. [Crossref] [PubMed]
- Sibley CR, Blazquez L, Ule J. Lessons from non-canonical splicing. Nat Rev Genet 2016;17:407-21. [Crossref] [PubMed]
- Zhang PF, Pei X, Li KS, et al. Circular RNA circFGFR1 promotes progression and anti-PD-1 resistance by sponging miR-381-3p in non-small cell lung cancer cells. Mol Cancer 2019;18:179. [Crossref] [PubMed]
- Sun M, Zhao W, Chen Z, et al. Circular RNA CEP128 promotes bladder cancer progression by regulating Mir-145-5p/Myd88 via MAPK signaling pathway. Int J Cancer 2019;145:2170-81. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384-8. [Crossref] [PubMed]
- Wang J, Jiang C, Li N, et al. The circEPSTI1/mir-942-5p/LTBP2 axis regulates the progression of OSCC in the background of OSF via EMT and the PI3K/Akt/mTOR pathway. Cell Death Dis 2020;11:682. [Crossref] [PubMed]
- Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 2012;13:271-82. [Crossref] [PubMed]
- Li S, Yan G, Liu W, et al. Circ0106714 inhibits tumorigenesis of colorectal cancer by sponging miR-942-5p and releasing DLG2 via Hippo-YAP signaling. Mol Carcinog 2020;59:1323-42. [Crossref] [PubMed]
- Wang Q, Wu J, Huang H, et al. lncRNA LIFR-AS1 suppresses invasion and metastasis of non-small cell lung cancer via the miR-942-5p/ZNF471 axis. Cancer Cell Int 2020;20:180. [Crossref] [PubMed]
- Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol 2017;14:611-29. [Crossref] [PubMed]
- Stevens TA, Meech R. BARX2 and estrogen receptor-alpha (ESR1) coordinately regulate the production of alternatively spliced ESR1 isoforms and control breast cancer cell growth and invasion. Oncogene 2006;25:5426-35. [Crossref] [PubMed]
- Sellar GC, Li L, Watt KP, et al. BARX2 induces cadherin 6 expression and is a functional suppressor of ovarian cancer progression. Cancer Res 2001;61:6977-81. [PubMed]


