
Whole exome sequencing characterization of individuals presenting extreme phenotypes of high and low risk of developing tobacco-induced lung adenocarcinoma
Introduction
Lung neoplasms account for the majority of cancer deaths worldwide (1), and their development is strongly linked to tobacco (2,3). Yet, despite the robustness of this association, some heavy smokers develop lung cancer at young ages, while others do not develop it at advanced ages, suggesting that large interindividual differences in the susceptibility to develop tobacco-induced lung cancer exist. The characterization of the potential molecular causes underlying these characteristic phenotypes could allow high-risk populations, that could benefit from prevention and screening programs, to be identified. Most importantly, it could lead to the discovery of the biological mechanisms that explain the existence of individuals presenting phenotypes of increased and decreased susceptibility to developing this disease.
Here, we report for the first time the results of a whole exome sequencing (WES) study of the germline DNA of individuals presenting clinical phenotypes of very high and very low risk of developing tobacco-induced lung adenocarcinoma. For this purpose, we selected individuals who were heavy smokers and either developed lung adenocarcinoma at an early age; or did not develop lung adenocarcinoma -as well as any other tumors, related or unrelated to tobacco- at an advanced age. We aimed to identify new susceptibility variants associated with these clinically relevant phenotypes. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tlcr-20-1197).
Methods
Study design
We performed an extreme phenotype study, with the aim of increasing the efficiency of discovering genetic alterations associated with the risk of developing lung adenocarcinoma induced by tobacco. We hypothesized that risk-related genetic alterations would be enriched in the phenotypic extremes, and consequently, a careful selection of individuals presenting the extreme phenotypes of interest would facilitate the identification of such alterations (4-6). This methodology has allowed highly relevant cancer biomarkers to be identified, as reviewed elsewhere (7,8).
The cancer cohort subjects (extreme cases) were selected from heavy smokers (≥15 pack-years) presenting a histologically confirmed diagnosis of lung adenocarcinoma at an early age (≤55 years). We also included one patient who developed lung adenocarcinoma at an extremely young age (37 years) and presented tobacco consumption below 15 pack-years (6), given his phenotypic relevance, and because we assumed that he was too young to achieve the selected threshold of tobacco consumption. Patients with known driver genetic alterations susceptible of targeted therapy (EGFR, ALK, ROS1, BRAF genetic alterations, etc.) were not included in the study, in order to increase the molecular homogeneity of the selected individuals.
The cancer-free cohort individuals (extreme controls) were selected from heavy smokers (≥15 pack-years) who had not developed lung adenocarcinoma -or any other cancer, related or not to tobacco- at an advanced age (≥72 years).
The thresholds for age and tobacco consumption were set in order to identify from our series those individuals presenting the most extreme phenotypes regarding the individual risk of developing tobacco-induced lung adenocarcinoma.
Extreme cases and controls were recruited among 3,631 subjects included in the databases of the Center for Applied Medical Research (CIMA, Pamplona, Spain) and the University Clinic of Navarra (Pamplona, Spain), from the University of Navarra.
Samples and data from patients included in the study were provided by the Biobank of the University of Navarra and were processed following standard operating procedures approved by the Ethics and Scientific Committees.
DNA extraction and genotyping by WES
Genomic DNA was obtained from peripheral EDTA-blood using the QIAamp DNA Mini Kit (Qiagen Iberia, Madrid, Spain) following the manufacturer’s instructions, and were stored at −20 °C until use. Genotyping was performed with a low input protocol using 800 ng of germline DNA after analysis in a TapeStation system (Agilent, Santa Clara, USA) and sequencing with the Agilent Human Exome Capture v5 kit at 2×100 bp and medium coverage at 75× in a NovaSeq system (Illumina, San Diego, USA). All exome data files are available from the authors upon reasonable request.
Statistical analysis
After quality control (FastQC) and trimming of the reads (trimmomatic) (9), we performed the read alignment using a BWA-MEM aligner and the hg38 human assembly as a reference (10). The resulting BAM files were processed using an analysis pipeline of variant calling based on the GATK best practices (11). Several filters were applied such us the variant score normalized by allele depth for a variant (DP <20), the root mean square of the mapping quality of reads across all samples and the strand biases estimated by both Fisher’s Exact Test (FS >60) and Symmetric Odds Ratio Test (SOR >40) among others.
The detected mutations were then annotated using ANNOVAR with different databases of genome localization, variant effect prediction, population SNPs (ExAC and 1000genomes) and clinical association of variants (ACMG, ClinVar, dbSNP and COSMIC) (12). All the results (point mutations and indels) were integrated and analyzed using statistical methods in R/Bioconductor (13). We selected nonsynonymous variants located in exonic regions and in the intronic splice site flanking regions. Types of selected variants included nonsynonymous single nucleotide variants (SNVs), frameshift insertions and deletions, non-frameshift insertions and deletions and stop gains and losses. The comparison of allele frequencies between the experimental groups was performed using “allelic” R package (14) and the plots were developed with “ComplexHeatmap” R package (15). In the case of genes harboring 3 or more variants that showed a significant difference in their allele frequencies (P<0.05), a burden analysis at the gene level was performed using “REBET” R package (16).
Validation of results of extreme cases using data from The Cancer Genome Atlas (TCGA)
We downloaded WES germline data from lung cancer patients included in TCGA (17) from the dbGaP database (https://www.ncbi.nlm.nih.gov/gap/), and we selected all the available patients presenting phenotypes similar to our extreme controls, i.e., heavy smokers (≥15 pack-years) diagnosed with lung adenocarcinoma at a young age (≤55 years). In these TCGA patients we assessed the germline variants that were significantly different between our extreme cases and controls, and we compared these data with our study groups.
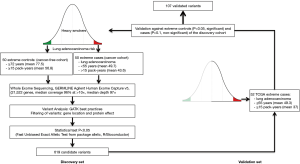
Data processing and statistical were similar to the methodology applied in our study groups and have been described in the previous section. We considered that a given variant was validated if it maintained statistical significance (P<0.05) when extreme TCGA cases were compared to our extreme controls, while being non-significant (P>0.1) in the comparison against our extreme cases (Figure 1).
We explored the diseases and biological pathways related to the genes harboring the variants that were validated using the Reactome Pathway Database (18) and Ingenuity Pathway Analysis (https://www.qiagenbioinformatics.com), which include manually curated and fully traceable data derived from literature sources.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Ethics Board of Clinical Universidad de Navarra (reference number 165/2015) and informed consent was obtained from all the patients.
Results
We performed germline WES in 100 Caucasian individuals presenting extreme phenotypes of very high and very low risk of developing tobacco-induced lung adenocarcinoma, 50 extreme cases and 50 extreme controls (Figure 1). The mean age for the cancer and cancer-free cohorts was respectively 49.7 (range, 34–55) and 77.5 years (range, 72–90). Mean tobacco consumption was respectively 43.6 (range, 6–129.5) and 56.8 pack-years (range, 20–123.8). Additional characteristics are displayed on Table S1.
Median exome sequencing coverage was 96% at >10× and median depth was 97×. We identified 619 variants that were differentially represented in their genotype frequencies (P<0.05) between both cohorts, located in 475 genes in Supplementary online file (https://cdn.amegroups.cn/static/application/3d42322f0a2368cd1833caaa0d1ccb48/tlcr-20-1197-1.xlsx).
The most significant variants were located in ALPK2 (P=4.48×10−5), HLA-A (2 variants, presenting respectively P=7.68×10−5 and P=1.53×10−4) and CRIPAK (P=2.24×10−4). The 50 most significant variants are represented on Figure 2.
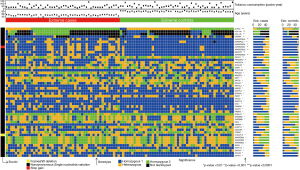
Twenty-three genes included ≥3 significantly different genetic variants (range, 3–11, Table S2). The genes harboring more significant variants were PRAMEF2 (11 variants) and GBP4 (8 variants). Among the genes harboring more than three significant variants, those obtaining the most significant P values at the gene level were ANKRD36C (P=2.28×10−14) and PRAMEF2 (P=1.2×10−12) (Table S2).
Validation of results using extreme cases obtained from TCGA
We validated our results in all the individuals available from TCGA who presented the same clinical characteristics as our extreme cases, i.e., heavy smokers (≥15 pack-years) presenting a histologically confirmed diagnosis of lung adenocarcinoma at an early age (≤55 years), according to the established criteria (Figure 1). We identified 52 extreme cases, who presented a mean age of 49.3 years (range, 33–55) and a mean tobacco consumption of 37 pack-years (range, 15–120).
From the 619 variants previously identified, 547 were assessable in the TCGA germline data, 108 of which were validated, according to the previously stated criteria: statistical significance (P<0.05) when extreme TCGA cases were compared to our extreme controls, while being non-significant (P>0.1) in the comparison against our extreme cases. All the validated variants are presented in Table S3.
Table 1 represents the variants that obtained the most significant P values in the validation study (19-24). These variants achieved statistical significance in the False Discovery Rate (FDR) test for the comparison of TCGA extreme cases with our extreme controls. From all the validated variants, those presenting the highest significance in the identification study were located in ALPK2 and HNRNPCL1.

Full table
Three genes harbored ≥3 validated variants: PRAMEF19 (6 variants), RFPL2 (5 variants) and PRAMEF2 (3 variants). Genes harboring more than 1 validated variants are presented in Table S4.
We explored the diseases related to the genes harboring the validated variants using Ingenuity Pathway Analysis. Forty-eight of the most significant 50 disease categories obtained for such genes were related to cancer, with P values ranging from 2.18×10−9 to 3.88×10−5 (data not shown).
We performed a Reactome Pathway Database analysis which indicated that the most significant biological pathways associated with the genes harboring the validated variants were related to immune regulation, interferon and cytokine signaling, and antigen processing and presentation, achieving significant P values in the global analysis and in the FDR test (Table 2).

Full table
Discussion
In this study we report for the first-time novel germline variants associated with individuals presenting extreme phenotypes of very high and very low risk for developing lung adenocarcinoma induced by tobacco, assessed by WES. We validated our results in an independent group including all the available patients from TCGA who presented the same clinical characteristics as our extreme cases. Many of the most significant validated variants belong to genes associated with relevant pathways related to cancer initiation and development, such as ALPK2, HLA-A, PARP4 or NQO1, among others. The genes harboring the validated variants were markedly associated with cancer disease categories, according to the Ingenuity Pathway Analysis; and they were strongly related to antigen presentation and immune regulation pathways, according to the Reactome Pathway Analysis.
Tobacco-induced lung cancer is one of the most relevant challenges to public health. The identification of molecular factors associated with either increased risk, or intrinsic protection from developing this disease, could allow to identify high-risk populations, in which tobacco cessation and screening programs may be most beneficial. Moreover, it could improve our understanding of the mechanisms of carcinogenesis and physiological protection against developing cancer, which may guide new approaches. Remarkably, although our study was specifically designed for lung adenocarcinoma, the individuals in the cancer-free cohort did not develop any other tumors. Consequently, our results may also be relevant to other neoplasms, especially those related to tobacco.
ALPK2, mapped to 18q21.31, is a tumor suppressor gene regulated by oncogenic KRAS. (25) The dependence on KRAS is of particular relevance, since lung adenocarcinoma frequently presents KRAS mutations (26). ALPK2 is crucial for luminal apoptosis in normal colonic crypts and plays a critical role in the up-regulation of DNA repair genes in the normal colonic epithelium, including RAD51, the Fanconi anemia complementation genes FANCA, FANCE and FANCG and the Bloom syndrome gene (BLM) (25). ALPK2 is down-regulated in human colonic adenomas, as compared to normal colonic mucosa, thus suggesting its involvement in early neoplastic transformation (25). Lawrence et al. found that ALPK2 was a novel mutated gene in human cancers in a large-scale genomic analysis of 4,742 human neoplasms and their matched normal tissue samples (27). In mice xenograft models, knockdown of ALPK2 inhibits the development and progression of ovarian cancer (28) and renal cell cancer (29), thus supporting its relevance in cancer initiation and development.
Interestingly, four of the variants identified involve the alpha1 and alpha2 domains of HLA-A molecules in areas oriented to the antigenic peptide binding cleft. HLA-A is a highly polymorphic locus incorporating over 450 allelic protein variants. The amino acid sequence variants discovered in this study are shared by different HLA-A allele subgroups generating diallelic polymorphisms that could be important for antigen presentation to CD8 T-cells and thereby could play a role in immune surveillance against cancer. This is especially relevant in tumors induced by carcinogens, (i.e., tobacco) potentially giving rise to multiple neoantigens. The aminoacid sequences found to be protective in homozygosis are shared by multiple HLA-A alleles, but at least three of the protective aminoacids in positions 86, 101 and 151 are shared by alleles HLA-A23 and HLA-A24. No in-depth studies exist on the correlation between cancer susceptibility and HLA-A allelic variants, and these findings warrant detailed analysis to address the potential influence of such polymorphisms on immune surveillance.
We validated additional variants in genes related to functions that are relevant for cancer initiation and development (Table 1). Moreover, an Ingenuity Pathway Analysis of the genes harboring the validated variants revealed that they are strongly linked to cancer-related diseases; and the Reactome Pathway Database analysis showed that their significant association to immune regulation, interferon and cytokine signaling and antigen processing and presentation. These findings support the notion that the variants identified and validated in our study are linked to genes that are markedly associated with cancer development, and consequently, they represent appealing targets for future research.
Increased risk of developing cancer has been firmly linked to specific genetic alterations, such as BRCA1/2 (30) or TP53 mutations (31,32), among others. These and other similar alterations were discovered through the identification of individuals presenting cancer at early ages and/or with familial aggregation. Here, we have followed a similar methodology to define the genetic alterations related with high risk of developing lung adenocarcinoma, maintaining age as a selection factor and substituting familial aggregation for cancer presentation following heavy exposure to a well-known risk factor, such as tobacco.
To our knowledge, no genetic alteration has ever been confirmed to induce protection against cancer development in human beings. Nevertheless, protective genetic alterations have been described in other fields of Medicine. For example, alterations in the chemokine coreceptor CCR5 confer complete protection against certain strains of the Human Immunodeficiency Virus (HIV) (33,34); APOE3 homozygote mutations have been reported to confer protection against autosomal dominant Alzheimer’s disease (35); and nonsense mutations in PCSK9, are associated with low levels of serum low density lipoproteins (LDL) and low risk of cardiovascular disease (36). Remarkably, the first two examples were discovered through the study of individuals highly exposed to well-known risk factors for developing the disease under study, i.e., high exposure to HIV-1 (37); and presence of presenilin 1 (PSEN1) mutations, which are associated with autosomal dominant Alzheimer disease (38). As for PCSK9 mutations, they were discovered through the study of individuals presenting extreme phenotypes consisting of very low serum LDL levels. The success of these approaches supports the methodology that we have developed for the present study.
In a previous study we applied the same methodology, studying as extreme cases individuals with non-small lung cancer, rather than exclusively adenocarcinoma; and assessing the germline DNA through Genome Wide Association Study (GWAS) (39). We identified and validated two new genetic variants in ATP10D and PDE10A which were differentially expressed in individuals presenting the extreme phenotypes assessed, and we confirmed the prognostic relevance of the associated proteins in early stage non-small cell lung cancer. We defined that protected individuals presented proficient cancer-risk phenotypes (PROCARPs) and high-risk individuals showed deficient cancer-risk phenotypes (DECARPs). For the present study, we focused exclusively on patients presenting lung adenocarcinoma, in order to increase the homogeneity of the phenotypes studied; and we used WES to maximize the chance of identifying gene variants that impact protein synthesis.
The main limitation of our study is that, despite our efforts to recruit highly selected individuals, phenotypic heterogeneity may persist from a clinical, pathological and molecular standpoint. Regarding clinical variables, phenotype selection may be improved by including individuals presenting even more extreme ages and increased tobacco exposure (i.e., higher tobacco consumption and/or shorter tobacco cessation intervals, active smokers, …). Additional inclusion criteria could increase homogeneity, such as presenting a similar stage of the disease (i.e., either early or advanced tumors). From a pathological and molecular perspective, heterogeneity could be further reduced by analyzing patients presenting exclusively specific lung carcinoma subtypes, and/or specific tumor molecular profiles, such as either KRAS positive or negative tumors, as well as other molecular alterations. These and other variables were not controlled for in our design because of sample size limitations. We hypothesize that further ongoing efforts to increase the sample size while improving the selection of extreme phenotypes will maximize the power of this strategy. Demographical variables, such as the ethnic background should also be taken into consideration in future studies. In this sense, there are emerging initiatives for developing population-specific genetic databases that can address the local genetic component and the population stratification level, such as the Collaborative Spanish Variability Server (CSVS) (40). Due to sample availability, the individuals included in our study were mainly of Caucasian origin, which may limit the applicability of our findings.
An additional limitation is the possibility that extreme controls may develop lung adenocarcinoma in the future. Yet, this limitation is controlled for by the large differences between the mean ages of both groups, which indicate at least an appreciable delay in the development of clinically assessable tumors. Also, the unlikely possibility that lung adenocarcinoma may have been induced by causes other than tobacco in some extreme cases, must be considered. In addition, while the relevant literature supports the relevance in cancer development of the genes that we validated, the biological effects associated with the variants that we report must be established in functional studies. Importantly, our study reveals which variants were overrepresented in each group of individuals, but it must be determined whether their effects may be associated either with the increased risk observed in cases, or with the increased protection observed in controls. Indeed, additional research is needed to further develop this methodology, including the optimal definition of extreme phenotypes, the determination of the sample size, or the extrapolation of the results to general patient populations (e.g., extreme phenotype vs. control population designs) (41).
Conclusions
In summary we have characterized for the first time with WES the germline background of individuals presenting extreme phenotypes of very high and very low risk for developing tobacco-induced lung adenocarcinoma. Our findings may allow individuals presenting high and low risk of developing this tumor to be identified and the molecular mechanisms that explain these clinically relevant phenotypes to be characterized. Consequently, our results and our methodology warrant further development through the comprehensive molecular characterization with different techniques of larger groups of individuals presenting these well-defined and carefully selected extreme phenotypes.
Acknowledgments
We particularly acknowledge our patients for their participation and the Biobank of the University of Navarra for its collaboration. We are thankful to Dr. Paul Miller (University of Navarra) for review of the manuscript; and to Julián Sánchez-Ostiz Erviti for his support in management functions. The results published here are in part based upon data generated by the TCGA Research Network:
Presented in part at the 2018 Meeting of the European Society of Medical Oncology (ESMO); 2019 Meeting of the American Society of Clinical Oncology (ASCO); and 2020 Meeting of the Society for Immunotherapy of Cancer (SITC).
Funding: This work was supported by the Spanish Society of Medical Oncology; Fundación SEOM and Fundación Salud 2000; and Government of Navarra. LMM research work is supported by Foundation for Applied Medical Research (FIMA), Fundación Científica de la Asociación Española Contra el Cáncer, Fundación Ramón Areces, and Fondo de Investigación Sanitaria-Fondo Europeo de Desarrollo Regional “Una manera de hacer Europa” (PI19/00098).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-20-1197
Peer Review File: Available at http://dx.doi.org/10.21037/tlcr-20-1197
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-20-1197
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-20-1197). Dr. JLPG reports grants from Spanish Society of Medical Oncology, grants from Fundación SEOM and Fundación Salud 2000, grants from Government of Navarra, during the conduct of the study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Ethics Board of Clinical Universidad de Navarra (reference number 165/2015) and informed consent was obtained from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Hoffman FL. Cancer and Smoking Habits. Ann Surg 1931;93:50-67. [Crossref] [PubMed]
- Müller FH. Tabakmißbrauch und Lungencarcinom. Z Krebsforsch 1939;49:57-85. [Crossref]
- Perez-Gracia JL, Sanmamed MF, Bosch A, et al. Strategies to design clinical studies to identify predictive biomarkers in cancer research. Cancer Treat Rev 2017;53:79-97. [Crossref] [PubMed]
- Zhang G, Nebert DW, Chakraborty R, et al. Statistical power of association using the extreme discordant phenotype design. Pharmacogenet Genomics 2006;16:401-13. [Crossref] [PubMed]
- Amanat S, Requena T, Lopez-Escamez JA. A Systematic Review of Extreme Phenotype Strategies to Search for Rare Variants in Genetic Studies of Complex Disorders. Genes 2020;11:987. [Crossref] [PubMed]
- Perez-Gracia JL, Ruiz-Ilundain MG. Cancer protective mutations: Looking for the needle in the haystack. Clin Transl Oncol 2001;3:169-71.
- Perez-Gracia JL, Ruiz-Ilundain MG, Garcia-Ribas I, et al. The role of extreme phenotype selection studies in the identification of clinically relevant genotypes in cancer research. Cancer 2002;95:1605-10. [Crossref] [PubMed]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114-20. [Crossref] [PubMed]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754-60. [Crossref] [PubMed]
- Van der Auwera GA, Carneiro MO, Hartl C, et al. From fastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Curr Protoc Bioinforma 2013;43:11.10.1-11.10.33.
- Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164 [Crossref] [PubMed]
- R: The R Project for Statistical Computing. 2020. Available online: https://www.r-project.org/
- Guedj M, Wojcik J, Della-Chiesa E, et al. A fast, unbiased and exact allelic test for case-control association studies. Hum Hered 2006;61:210-21. [Crossref] [PubMed]
- Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:2847-9. [Crossref] [PubMed]
- Zhu B, Mirabello L, Chatterjee N. A subregion-based burden test for simultaneous identification of susceptibility loci and subregions within. Genet Epidemiol 2018;42:673-83. [Crossref] [PubMed]
The Cancer Genome Atlas Program - National Cancer Institute Reactome Pathway Database 2020 . Available online: https://reactome.org/- Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPs, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci 2008;13:3046-82. [Crossref] [PubMed]
- Li L, Li Y, Bai Y. Role of GSDMB in Pyroptosis and Cancer. Cancer Manag Res. 2020;12:3033-43. [Crossref] [PubMed]
- Cassandri M, Smirnov A, Novelli F, et al. Zinc-finger proteins in health and disease. Cell Death Discov 2017;3:17071. [Crossref] [PubMed]
- Franchini M, Lippi G. The role of von Willebrand factor in hemorrhagic and thrombotic disorders. Crit Rev Clin Lab Sci 2007;44:115-49. [Crossref] [PubMed]
- Funauchi Y, Tanikawa C, Yi Lo PH, et al. Regulation of iron homeostasis by the p53-ISCU pathway. Sci Rep 2015;5:16497. [Crossref] [PubMed]
- Zhang K, Chen D, Ma K, et al. NAD(P)H:Quinone Oxidoreductase 1 (NQO1) as a Therapeutic and Diagnostic Target in Cancer. J Med Chem 2018;61:6983-7003. [Crossref] [PubMed]
- Yoshida Y, Tsunoda T, Doi K, et al. ALPK2 is Crucial for luminal apoptosis and DNA repair-related gene expression in a three-dimensional colonic-crypt model. Anticancer Res 2012;32:2301-8. [PubMed]
- Rodenhuis S, van de Wetering ML, Mooi WJ, et al. Mutational activation of the K-ras oncogene. A possible pathogenetic factor in adenocarcinoma of the lung. N Engl J Med 1987;317:929-35. [Crossref] [PubMed]
- Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014;505:495-501. [Crossref] [PubMed]
- Zhu X, Yan S, Xiao S, et al. Knockdown of ALPK2 inhibits the development and progression of Ovarian Cancer. Cancer Cell Int 2020;20:267. [Crossref] [PubMed]
- Jiang J, Han P, Qian J, et al. Knockdown of ALPK2 blocks development and progression of renal cell carcinoma. Exp Cell Res 2020;392:112029 [Crossref] [PubMed]
- Hall JM, Lee MK, Newman B, et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science 1990;250:1684-9. [Crossref] [PubMed]
- Miller RW. Deaths from childhood cancer in sibs. N Engl J Med 1968;279:122-6. [Crossref] [PubMed]
- Li FP, Fraumeni JF. Rhabdomyosarcoma in children: Epidemiologic study and identification of a familial cancer syndrome. J Natl Cancer Inst 1969;43:1365-73. [PubMed]
- Liu R, Paxton WA, Choe S, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 1996;86:367-77. [Crossref] [PubMed]
- Quillent C, Oberlin E, Braun J, et al. HIV-1-resistance phenotype conferred by combination of two separate inherited mutations of CCR5 gene. Lancet 1998;351:14-8. [Crossref] [PubMed]
- Arboleda-Velasquez JF, Lopera F, O’Hare M, et al. Resistance to autosomal dominant Alzheimer’s disease in an APOE3 Christchurch homozygote: a case report. Nat Med 2019;25:1680-3. [Crossref] [PubMed]
- Cohen J, Pertsemlidis A, Kotowski IK, et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet 2005;37:161-5. [Crossref] [PubMed]
- Rowland-Jones S, Sutton J, Ariyoshi K, et al. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat Med 1995;1:59-64. [Crossref] [PubMed]
- Lopera F, Ardilla A, Martinez A, et al. Clinical features of early-onset Alzheimer disease in a large kindred with an E280A presenilis-1 mutation. Am J Ophthalmol 1997;124:137-8. [Crossref]
- Fusco JP, Pita G, Pajares MJ, et al. Genomic characterization of individuals presenting extreme phenotypes of high and low risk to develop tobacco-induced lung cancer. Cancer Med 2018;7:3474-83. [Crossref] [PubMed]
- Peña-Chilet M, Roldán G, Perez-Florido J, et al. CSVS, a crowdsourcing database of the Spanish population genetic variability. Nucleic Acids Res 2021;49:D1130-7. [Crossref] [PubMed]
- Emond MJ, Louie T, Emerson J, et al. Exome Sequencing of Phenotypic Extremes Identifies CAV2 and TMC6 as Interacting Modifiers of Chronic Pseudomonas aeruginosa Infection in Cystic Fibrosis. PLoS Genet 2015;11:e1005273 [Crossref] [PubMed]


