
Earlier extracranial progression and shorter survival in ALK-rearranged lung cancer with positive liquid rebiopsies
Introduction
“Liquid biopsies” analyzing cell-free genetic material of the tumor in the patients’ blood (Circulating tumor DNA; ctDNA) have emerged as a non-invasive method for the molecular profiling of various malignancies (1,2). Non-small-cell lung cancer (NSCLC), and in particular anaplastic lymphoma kinase-rearranged (ALK+) tumors are emerging as a model disease in this regard (3,4). ALK+ NSCLC patients can experience long-lasting responses under tyrosine kinase inhibitors (TKI) (5-7), with treatment failure frequently occurring due to resistance mutations in exons coding for the ALK kinase domain (8-10). Several proof-of-principle studies have recently shown that mechanisms of acquired resistance to ALK inhibitors can be efficiently captured by longitudinal ctDNA assays, which therefore represent a promising tool to guide the selection of tailored next-line therapies (11-16). The current study focusses on how the potential clinical utility of liquid rebiopsies in ALK+ NSCLC depends on the anatomical pattern of treatment failure, and on the prognostic information that their results additionally provide about the subsequent disease course and patient survival. We present this article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tlcr-21-32).
Methods
Study population, objectives, collection of clinical data, ethics
This study included all ALK+ NSCLC patients treated with TKI between 2014 and 2019 in our institution with ctDNA results at the time of radiologic changes. There were two main study objectives: (I) relationship between the results of liquid rebiopsies and the anatomic pattern of disease progression; and (II) the survival [i.e., progression-free survival (PFS), time-to-next treatment and overall survival (OS)] of progressive patients according to the results of liquid rebiopsies. Blood samples had been prospectively collected from each patient at each visit every 8–12 weeks during the entire disease course, as previously published (17). Clinical data were systematically collected from the medical records with a cut-off in June 2020. There were no lost-to-follow-up cases, as all patients were treated in-house. In order to control for possible confounders, every case was annotated regarding baseline clinical characteristics and treatment in great detail, and these data were analyzed together with parameters of interest, including multivariable testing. Particular attention was given to the localization of disease progression, i.e., intra- vs. extracranial, and intra- vs. extrathoracic, as well as to parameters that could influence patient outcome, i.e., the ECOG performance status (PS), molecular characteristics as outlined in the next section, whether the ongoing systemic treatment was switched at the time of liquid biopsy or continued beyond disease progression, the type of TKI treatment in conjunction with molecular results, and the administration of local therapies in case of oligoprogression. For PFS, the progression date documented in the records was verified by the investigators with review of radiologic images, i.e., chest/abdomen CT and brain MRI-based restaging every 6–12 weeks, without formal RECIST reevaluation, as several studies have demonstrated very good agreement between real-world and RECIST-based assessments (18,19). For evaluating the potential clinical utility of liquid biopsies in case of anatomically restricted disease progression and continuation of the same systemic treatment, changes in tumor size were considered regardless of the RECIST threshold, because any significant increase of tumor lesions poses therapeutic dilemmas to the treating physicians, and no uniform definition of oligoprogression exists in the literature (20). Time-to-next-treatment (TNT) was calculated from the start of the respective treatment line until change of systemic therapy or death. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the ethics committee of the Heidelberg University (S-296/2016). Written informed consent was provided by every participant.
Molecular analyses
Diagnosis of NSCLC was performed at the Institute of Pathology Heidelberg from tissue specimens according to the criteria of the current WHO Classification (2015) for lung cancer (21). Newly diagnosed cases were screened for the presence of an ALK alteration by fluorescence in situ hybridization (FISH, ZytoLight SPEC ALK probe, ZytoVision GmbH, Bremerhaven, Germany) and reverse-transcription polymerase chain reaction until 2015, or by immunohistochemistry (D5F3 clone, Roche, Mannheim, Germany) and RNA-based next-generation sequencing (NGS, ThermoFisher Lung Cancer Fusion Panel, Waltham, MA, USA) thereafter, as previously described (22). Molecular workup at baseline also included DNA-based NGS covering all exons of EGFR as well as several other genes with potentially actionable alterations (e.g., BRAF, MET, HER2) and/or considered relevant for lung cancer biology (e.g., exons 4–10 of TP53), as published (23). For liquid biopsies, cfDNA was isolated from 2 mL plasma using the AVENIO cfDNA Isolation Kit (Roche Diagnostics, Mannheim, Germany) and quantified with the Qubit dsDNA High Sensitivity Kit (ThermoFisher). Enrichment of the characteristic mononucleosomal fragment peak (160–200 bp) and absence of contaminating high molecular weight genomic DNA (24,25) were verified using the Bioanalyzer 2100 High Sensitivity DNA Kit (Agilent Technologies, Santa Clara, CA, USA). Sequencing libraries were constructed from 1.5–50 ng cfDNA (median 20.3 ng), using the AVENIO ctDNA Library Preparation Kit with the AVENIO Targeted and the AVENIO Surveillance Panel (both from Roche), which enrich a 17-gene (81 kb) and 197-gene (198 kb) target region, respectively, as described previously (16). Only genomic regions covered by both panels were considered for the data reported here, i.e., fusions in ALK intron 19, RET intron 11, ROS1 introns 31–15; single nucleotide variants (SNV) in ALK exons 3–7/9/10/12/14/15/16/19–28, all exons of TP53, KRAS, EGFR, MET, ERBB2, BRCA1/2, selected exons of BRAF [3/8/10-18], NRAS [2–4], APC [2/3/5–7/9/10/12–16], KIT [2/3/5/8–20], PDGFRA [3–23], RET [2/5/7/9/10/12–19], ROS1 [10/15/16/22/23/27/36–42], as well as copy number alterations in MET, EGFR and ERBB2. Equimolar amounts of 16 libraries were pooled and sequenced on an Illumina NextSeq550 using the High Output Kit V2 (300 cycles) according to the manufacturer’s protocol (Illumina, San Diego, CA, USA). Automated raw data processing and data analysis was performed with the AVENIO ctDNA analysis software (Roche, version 2.0.0), which applies the bioinformatics pipeline from CAPP-Seq (26) with integrated digital error suppression (27). The variant allele frequency cut-off for a positive SNV call was 0.01%. Called variants were verified manually for sufficient coverage (i.e., ≥2,500 read counts) and visualized using the Integrative Genomics Viewer (IGV) (28) as published (16).
Statistical methods
Survival data were analyzed according to Kaplan-Meier and compared between patient groups with the logrank test. The relative effects of liquid biopsy results and other clinical or molecular parameters on patient survival were analyzed with multivariable testing using a Cox regression model that included the liquid biopsy result (positive or negative), type of EML4-ALK variant, baseline TP53 status, baseline ECOG PS, the number of treatment lines before liquid biopsy, and whether the treatment was switched or continued beyond progression after the liquid biopsy. Since primary endpoint of our study was the relationship between ctDNA results and radiologic changes, samples from different progression time-points of the same patient were analyzed as independent, in order to account for the fact that several factors can influence liquid biopsy positivity differently in each sample of the same patient (e.g., progression site, progression rate, tumor volume, preceding treatment), and that these factors act similarly across different patients. Duration of follow-up was calculated using the reverse Kaplan-Meier method (29). Categorical data were compared with the chi-square, and numerical data with the Student’s t-test. Confidence intervals for proportions were computed according to Clopper-Pearson (30). Statistical calculations were performed with SPSS version 24 (IBM Corp., Armonk, NY, USA) and plots generated with GraphPad Prism version 7 (GraphPad Software, La Jolla, CA, USA). All statistical tests were two-sided. P values <0.05 were considered statistically significant. The oncoprint plot (Figure S1) was generated using the cBioPortal (31,32).
Results
Cohort overview
Overall, in 56 TKI-treated ALK+ NSCLC patients 139 instances of radiologic changes at the time of ctDNA profiling were identified, of which 133 corresponded to disease progression. The patient subsets and clinical characteristics are summarized in Figure 1 and Table 1. The number of samples drawn at the time of radiologic changes varied per patient according to the clinical course, with a mean value of 2.46 (range, 1–7) for the entire cohort. An overview of all identified mutations alongside other patient characteristics is shown in Figures S1,S2 and Table S1. Mean coverage for ctDNA NGS was 14,730× and 4,385× pre- and post-deduplication, respectively. In 6/6 cases with a suspected benign cause for the recent radiologic changes, namely pneumonitis or lung infection based on the patients’ history, clinical presentation and laboratory findings, the liquid biopsies were negative, and these were excluded from further analysis (Figure 1).
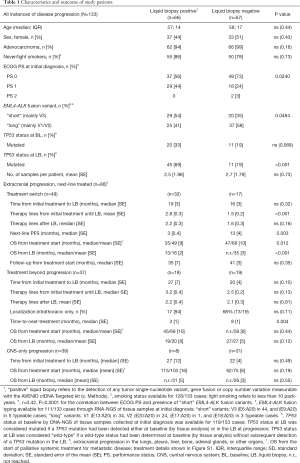
Full table
Liquid biopsy results according to the progression site
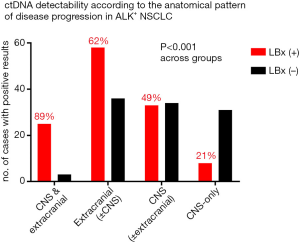
Among the remaining 133 cases, extracranial progression was noted in 71% (n=94), central nervous system (CNS) progression in 50% (n=67), and an isolated CNS progression in 29% of cases (n=39, Figure 1), with an overall percentage of liquid biopsy positivity at 50% (66/133, Table 1). The frequency of positive liquid biopsies was significantly higher with extracranial (62%) vs. isolated CNS progression (21%, P<0.001), and even higher with combined extra- and intracranial progression (89%, P<0.001, Figure 2). The infrequent cases with CNS-only progression and a positive liquid biopsy, tended to have a higher intracranial tumor load, i.e., multiple brain metastases including large lesions >1 cm in 8/8 patients (exemplary data shown in Figure S3). Apart from the higher intracranial tumor load, there was no other distinguishing radiologic characteristic of these cases, for example, radiologic evidence of meningeal carcinomatosis with isolated CNS progression was present in 2/8 cases with positive liquid biopsies vs. 2/31 cases with negative liquid biopsies. Baseline patient characteristics significantly associated with liquid biopsy positivity were presence of “short” echinoderm microtubule-associated protein-like 4 (EML4)-ALK fusion variants (mainly V3, in 54% vs. 35% of liquid biopsy-positive vs. negative cases, P=0.0484, Table 1) as detected by tissue RNA-NGS at initial diagnosis, and a worse ECOG PS at initial diagnosis (PS 0–1 in 56–44% vs. 73–24% of subsequently liquid biopsy-positive vs. negative cases, P=0.024, Table 1). The initial ECOG PS and presence of “short” EML4-ALK variants were positively correlated (r=0.42, P<0.001). In addition, presence of TP53 mutations at initial diagnosis as detected by tissue DNA-NGS, showed a trend for association with subsequent liquid biopsy positivity (baseline TP53 mutations in 33% vs. 19% of subsequently liquid biopsy-positive vs. negative cases, P=0.068, Table 1). The correlation between mutated TP53 status and liquid biopsy positivity became highly significant at the time of disease progression, when the TP53 results of liquid biopsies were also considered (69% vs. 19%, P<0.001, Table 1). Among patients with TP53 mutations detectable in tissue samples at baseline (n=13), some displayed TP53 mutations in subsequent ctDNA samples (n=6) and some not (n=7), while other patients tested TP53 wild-type at diagnosis and showed newly detectable TP53 mutations at progression (n=9, Figure S2).
Outcome of progressive patients according to liquid biopsy results
Next, we analyzed patient outcomes in conjunction with liquid biopsy results. OS for progressive patients was shorter in case of positive vs. negative liquid biopsies both from the time-point of liquid biopsy collection (13 vs. 28 months in median, P<0.001, Figure 3A), and from the start of palliative systemic treatment (58 vs. 67 months, P=0.005, Figure 3B). Subgroup analysis according to the site of disease progression, showed that this association between liquid biopsy results and OS pertained to cases with extracranial, but not with CNS-only progression (Figure 3C,D and Table 1).
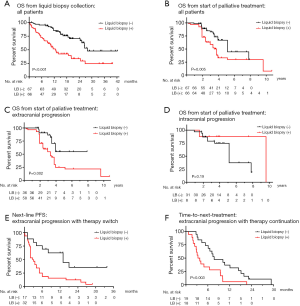
Within the subset of cases with extracranial disease progression and immediate switch to a different systemic treatment, the PFS of subsequent treatment line (3 vs. 13 months in median, P=0.003, Table 1 and Figure 3E), the OS from the time-point of liquid biopsy collection (13 months vs. not reached, P<0.001, Table 1), and the OS from start of palliative systemic treatment (35 vs. 47 months in median, P=0.012, Table 1) were shorter in case of positive compared to negative liquid biopsies. Similarly, within the subset with extracranial oligoprogression and continuation of the same systemic treatment, liquid biopsy positivity was associated with significantly shorter TNT (3 vs. 8 months, P=0.004, Figure 3F and Table 1). Of note, although liquid biopsies were drawn at comparable time-points after start of palliative systemic treatment in positive vs. negative cases (on average 19 vs. 16 months, P=0.32, Table 1 and Figure S1), cases with positive liquid biopsy results had already gone through significantly more therapy lines (mean 2.8 vs. 1.5, P<0.001, Table 1) and died earlier than cases with liquid biopsy negative results, despite the heavier treatment of the former. In multivariable analysis, liquid biopsy positivity in cases with extracranial progression was associated with shorter OS from the start of palliative systemic treatment, independent of other established predictors of worse outcome, i.e., presence of “short” ALK fusion variants, presence of TP53 mutations at diagnosis, initial ECOG PS, number of previous treatment lines, and treatment continuation or switch after the liquid biopsy (Table 2). The phenotype of liquid biopsy positivity at progression of ALK+ NSCLC according to the results of this study is summarized in Figure 4.

Full table
The alterations detected by liquid biopsies were sensitive to routinely available drugs in only 26% (17/66) of positive cases (Table S1). Detailed analysis of individual clinical courses showed that the great majority of cases (15/17 or 88%) had subsequently received ALK TKI with activity against the potentially druggable ALK mutations that were detected (Table S1).
Discussion
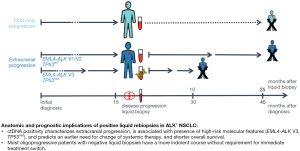
Main finding of the present study is the worse outcome of ALK+ NSCLC patients with positive liquid biopsies at the time of disease progression (Figure 4). At first, this might appear counterintuitive, because liquid rebiopsies are mainly employed to detect actionable genetic alterations and facilitate use of effective next-line targeted therapies (12). However, our results suggest that in most cases ctDNA detectability does do not go hand-in-hand with better therapeutic options. Even with the relatively narrow profiling focused on mutations of oncogenic drivers employed in this study, therapeutic opportunities emerged for less than half of liquid biopsy-positive cases, while genetic changes not sensitive to currently available therapies were more frequent (Figure S1 and Table S1). Moreover, ctDNA detectability correlated with presence of the high-risk molecular features “short” EML4-ALK fusion variant (mainly variant 3) and TP53 mutations, which were independently associated with worse patient outcome in several retrospective analyses and the randomized phase 3 trial ALTA-1L (33-38). The association between presence of EML4-ALK V3 and detectability of ALK resistance mutations has also been noted by other investigators (39) and stands in apparent contradiction to the adverse phenotype of V3-positive patients (40). This dissonance is reconciled by the results of the present study, which show that treatable ALK resistance mutations constitute a minor only fraction of acquired genetic alterations associated by EML4-ALK V3 (Figure S1). Frequent are also coexistence of multiple ALK mutations, which are usually associated with resistance to currently available TKI (41), and newly acquired TP53 mutations at the time of disease progression, which impair prognosis to an extent comparable to that of TP53 mutations present already at baseline (42). In addition, copy number alterations captured across the genome by shallow whole genome sequencing also accumulate faster in patients with EML4-ALK V3 and/or TP53 mutations, and are similarly associated with worse outcome (16). Nevertheless, it should be noted that the adverse prognostic impact of positive liquid rebiopsies goes beyond that of classical molecular risk factors in ALK+ NSCLC: in multivariable analysis, liquid biopsy positivity was associated with shorter overall survival independently of the EML4-ALK variant and the baseline TP53 status. Moreover, this adverse implication of positive liquid rebiopsies was also independent of an impairment in the initial ECOG PS (Table 2), which is a recognized feature of the adverse V3 phenotype likely caused by more extensive metastatic spread already at diagnosis (43,44). This worse outcome of patients with positive liquid rebiopsies fits nicely with the adverse prognostic implications of ctDNA detectability already noted for oncogene-driven NSCLC at initial diagnosis (45-48), and under ongoing TKI treatment (48-50).
The partial only concordance between baseline tissue-based and subsequent ctDNA-based TP53 results (Figure S2) is an additional argument for a combined tissue/liquid biopsy approach to molecular profiling in ALK+ NSCLC, as considerable spatial and temporal heterogeneity has been described for TP53 and other mutations in lung cancer (51,52), while at a purely technical level liquid biopsies can both capture some tissue-NGS negative cases, as well as miss some positive ones (53).
Another important characteristic of liquid biopsy-positive patients in our study was faster progression through treatment lines (Figure 4), regardless of whether ongoing systemic treatment was switched immediately (Figure 3E) or continued beyond oligoprogression (Figure 3F). Conversely, most patients with anatomically restricted progression and negative liquid biopsies appear to have a more indolent course. If this finding is confirmed by other studies, it could prove very useful for the stratification of oligoprogressive ALK+ NSCLC patients in clinical practice: for negative cases, continuation of current TKI treatment with or without local therapies could be a reasonable option, and was associated with a TNT >6 months in the majority of our patients, even without ablative measures in several cases (Table 1 and Figure S1). On the other hand, for positive cases, the liquid biopsy results not only identify patients with more aggressive disease and need for earlier change of systemic therapy (Figure 3F), but may also guide selection of the most suitable next-line compound based on the exact alteration detected.
For patients with isolated CNS progression, ctDNA detectability was low in our study, approximately one-third of that with extracranial progression (21% vs. 62%, Figure 2). These results together with similar findings of other investigators across various oncogene-driven NSCLC (54), collectively show that the intracranial compartment is largely inaccessible for blood-based liquid biopsies. Consequently, the clinical utility of plasma ctDNA assays in the setting of CNS-only progression is limited, for example their results were not associated with survival in our study (Figure 3D). Analysis of cerebrospinal fluid is preferrable to that of blood for molecular profiling of primary and metastatic CNS tumors, as other investigators have noted (55-58). Notwithstanding, our results show that blood-based liquid biopsies can be positive in some cases with extensive brain involvement, including multiple brain metastases and large (>1 cm) lesions (Figure S3). As brain metastases are known to disrupt the blood brain barrier, cases with more and larger intracranial lesions can be reasonably assumed to have a more disrupted blood brain barrier, which could facilitate export of ctDNA in the circulation.
Main limitation of our work is its retrospective character, therefore its results warrant confirmation in a larger, prospective study. At the same time, the findings appeared to be consistent in various analyses, for example, the worse prognosis of liquid biopsy-positive patients was evident regardless of whether treatment was switched or continued beyond progression. Also, we tried to control potential confounders, for example potentially relevant parameters were systematically collected for all patients (Figure S1), compared for cases with liquid biopsy-positive and negative results (Table 1), and included in multivariable testing (Table 2). Overall, the findings of the present study argue for ctDNA detectability at progression as a feature of biologically more aggressive disease, which is in line with observations in the treatment-naive setting (45,47). However, the exact values of PFS, TNT and OS observed in our cohort, although typical for an ALK+ NSCLC cohort, are not directly generalizable, and will certainly also be affected by the availability of more effective targeted therapies, like upfront lorlatinib (59), in the future. Also, the favorable outcome of oligoprogressive patients with negative liquid biopsies warrants further investigation in a larger cohort.
In summary, positivity of blood-based liquid rebiopsies in ALK+ NSCLC appears to characterize extracranial progression and biologically more aggressive disease. These results reconcile the increased detection of ALK resistance mutations with other features of the high-risk clinical phenotype associated with EML4-ALK V3 (22,39), especially in case of concomitant TP53 comutations (33). Conversely, most oligoprogressive ALK+ NSCLC patients with negative liquid biopsies appear to have a more indolent course without need for immediate change of systemic treatment.
Acknowledgments
We thank Ingrid Heinzmann-Groth and Saskia Östringer of the Translational Research Unit of the Thoraxklinik Heidelberg for assistance with collection of patient samples. We thank Simon Ogrodnik for excellent technical assistance and the Genomics Core Facility of the German Cancer Research Center (DKFZ) for sequencing and technical support.
Funding: This study was supported by the German Center for Lung Research (DZL) and the German Cancer Consortium (DKTK). The funding source did not have any influence on the design, conduction, and report of the results for this study.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tlcr-21-32
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tlcr-21-32
Peer Review File: Available at http://dx.doi.org/10.21037/tlcr-21-32
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tlcr-21-32). PC reports grants and personal fees from Novartis, grants and personal fees from Roche, grants and personal fees from AstraZeneca, personal fees from Pfizer, grants and personal fees from Takeda, personal fees from Chugai, personal fees from Boehringer, outside the submitted work; SD reports personal fees from Roche, outside the submitted work; DK reports personal fees from Pfizer, personal fees from BMS, personal fees from AstraZeneca, outside the submitted work; AV reports personal fees from AstraZeneca, outside the submitted work; MR reports personal fees from Amgen, personal fees from AstraZeneca, personal fees from BMS, personal fees from Boehringer, personal fees from Lilly, personal fees from Merck, personal fees from MSD, personal fees from Novartis, personal fees from Pfizer, personal fees from Roche, personal fees from Samsung, outside the submitted work; AS reports personal fees from AstraZeneca, personal fees from Lilly, personal fees from Bayer, personal fees from BMS, personal fees from Illumina, personal fees from Janssen, personal fees from MSD, personal fees from Novartis, personal fees from Pfizer, personal fees from Roche, personal fees from Seattle Genomics, outside the submitted work; MT reports grants from AstraZeneca, personal fees from Lilly, personal fees from BMS, personal fees from MSD, personal fees from Novartis, personal fees from Roche, personal fees from Boehringer, personal fees from Celgene, personal fees from Takeda, personal fees from AbbVie, outside the submitted work; HS reports grants and personal fees from Roche, outside the submitted work. The other authors have no conflicts of interest to declare. AS serves as an unpaid editorial board member of Translational Lung Cancer Research from Sep 2019 to Sep 2021. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics committee of the Heidelberg University (S-296/2016), and written informed consent was obtained from all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Volckmar AL, Sultmann H, Riediger A, et al. A field guide for cancer diagnostics using cell-free DNA: From principles to practice and clinical applications. Genes Chromosomes Cancer 2018;57:123-39. [Crossref] [PubMed]
- Keller L, Belloum Y, Wikman H, et al. Clinical relevance of blood-based ctDNA analysis: mutation detection and beyond. Br J Cancer 2021;124:345-58. [Crossref] [PubMed]
- Gadgeel SM, Mok T, Peters S, et al. Phase II/III blood first assay screening trial (BFAST) in patients (pts) with treatment-naïve NSCLC: Initial results from the ALK+ cohort. Ann Oncol 2019;30:v918. [Crossref]
- Madison R, Schrock AB, Castellanos E, et al. Retrospective analysis of real-world data to determine clinical outcomes of patients with advanced non-small cell lung cancer following cell-free circulating tumor DNA genomic profiling. Lung Cancer 2020;148:69-78. [Crossref] [PubMed]
- Breadner D, Blanchette P, Shanmuganathan S, et al. Efficacy and safety of ALK inhibitors in ALK-rearranged non-small cell lung cancer: A systematic review and meta-analysis. Lung Cancer 2020;144:57-63. [Crossref] [PubMed]
- Camidge DR, Kim HR, Ahn MJ, et al. Brigatinib versus Crizotinib in ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2018;379:2027-39. [Crossref] [PubMed]
- Mok T, Camidge DR, Gadgeel SM, et al. Updated overall survival and final progression-free survival data for patients with treatment-naive advanced ALK-positive non-small-cell lung cancer in the ALEX study. Ann Oncol 2020;31:1056-64. [Crossref] [PubMed]
- Lin JJ, Riely GJ, Shaw AT. Targeting ALK: Precision Medicine Takes on Drug Resistance. Cancer Discov 2017;7:137-55. [Crossref] [PubMed]
- Yu Y, Ou Q, Wu X, et al. Concomitant resistance mechanisms to multiple tyrosine kinase inhibitors in ALK-positive non-small cell lung cancer. Lung Cancer 2019;127:19-24. [Crossref] [PubMed]
- Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov 2016;6:1118-33. [Crossref] [PubMed]
- Dietz S, Christopoulos P, Gu L, et al. Serial liquid biopsies for detection of treatment failure and profiling of resistance mechanisms in KLC1-ALK-rearranged lung cancer. Cold Spring Harb Mol Case Stud 2019;5:a004630 [Crossref] [PubMed]
- McCoach CE, Blakely CM, Banks KC, et al. Clinical Utility of Cell-Free DNA for the Detection of ALK Fusions and Genomic Mechanisms of ALK Inhibitor Resistance in Non-Small Cell Lung Cancer. Clin Cancer Res 2018;24:2758-70. [Crossref] [PubMed]
- Shaw AT, Solomon BJ, Besse B, et al. ALK Resistance Mutations and Efficacy of Lorlatinib in Advanced Anaplastic Lymphoma Kinase-Positive Non-Small-Cell Lung Cancer. J Clin Oncol 2019;37:1370-9. [Crossref] [PubMed]
- Dagogo-Jack I, Brannon AR, Ferris LA, et al. Tracking the Evolution of Resistance to ALK Tyrosine Kinase Inhibitors through Longitudinal Analysis of Circulating Tumor DNA. JCO Precis Oncol 2018;2018:PO.17.00160.
- Manicone M, Scaini MC, Rodriquenz MG, et al. Liquid biopsy for monitoring anaplastic lymphoma kinase inhibitors in non-small cell lung cancer: two cases compared. J Thorac Dis 2017;9:S1391-6. [Crossref] [PubMed]
- Dietz S, Christopoulos P, Yuan Z, et al. Longitudinal therapy monitoring of ALK-positive lung cancer by combined copy number and targeted mutation profiling of cell-free DNA. EBioMedicine 2020;62:103103 [Crossref] [PubMed]
- Wessels S, Muley T, Christopoulos P, et al. Comprehensive serial biobanking in advanced NSCLC: feasibility, challenges and perspectives. Transl Lung Cancer Res 2020;9:1000-14. [Crossref] [PubMed]
- Ma X, Nussbaum NC, Magee K, et al. Comparison of real-world response rate (rwRR) to RECIST-based response rate in patients with advanced non-small cell lung cancer (aNSCLC). Ann Oncol 2019;30:v651. [Crossref]
- Bartlett CH, Mardekian J, Cotter M, et al. Concordance of real world progression free survival (PFS) on endocrine therapy as first line treatment for metastatic breast cancer using electronic health record with proper quality control versus conventional PFS from a phase 3 trial. Cancer Res 2018; [Crossref]
- Rheinheimer S, Heussel CP, Mayer P, et al. Oligoprogressive Non-Small-Cell Lung Cancer under Treatment with PD-(L)1 Inhibitors. Cancers (Basel) 2020;12:1046. [Crossref] [PubMed]
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Christopoulos P, Endris V, Bozorgmehr F, et al. EML4‐ALK fusion variant V3 is a high‐risk feature conferring accelerated metastatic spread, early treatment failure and worse overall survival in ALK+ non‐small cell lung cancer. Int J Cancer 2018;142:2589-98. [Crossref] [PubMed]
- Volckmar AL, Leichsenring J, Kirchner M, et al. Combined targeted DNA and RNA sequencing of advanced NSCLC in routine molecular diagnostics: Analysis of the first 3,000 Heidelberg cases. Int J Cancer 2019;145:649-61. [Crossref] [PubMed]
- Koessler T, Paradiso V, Piscuoglio S, et al. Reliability of liquid biopsy analysis: an inter-laboratory comparison of circulating tumor DNA extraction and sequencing with different platforms. Lab Invest 2020;100:1475-84. [Crossref] [PubMed]
- Mansukhani S, Barber LJ, Kleftogiannis D, et al. Ultra-Sensitive Mutation Detection and Genome-Wide DNA Copy Number Reconstruction by Error-Corrected Circulating Tumor DNA Sequencing. Clin Chem 2018;64:1626-35. [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol 2016;34:547-55. [Crossref] [PubMed]
- Robinson JT, Thorvaldsdóttir H, Wenger AM, et al. Variant Review with the Integrative Genomics Viewer. Cancer Res 2017;77:e31-4. [Crossref] [PubMed]
- Schemper M, Smith TL. A note on quantifying follow-up in studies of failure time. Control Clin Trials 1996;17:343-6. [Crossref] [PubMed]
- Clopper CJ, Pearson ES. The Use of Confidence or Fiducial Limits Illustrated in the Case of the Binomial. Biometrika 1934;26:404-13. [Crossref]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Christopoulos P, Kirchner M, Bozorgmehr F, et al. Identification of a highly lethal V3+ TP53+ subset in ALK+ lung adenocarcinoma. Int J Cancer 2019;144:190-9. [Crossref] [PubMed]
- Camidge DR, Niu H, Kim HR, et al. Correlation of baseline molecular and clinical variables with ALK inhibitor efficacy in ALTA-1L. J Clin Oncol 2020;38:9517. [Crossref]
- Yang CY, Liao WY, Ho CC, et al. Association of Programmed Death-Ligand 1 Expression with Fusion Variants and Clinical Outcomes in Patients with Anaplastic Lymphoma Kinase-Positive Lung Adenocarcinoma Receiving Crizotinib. Oncologist 2020;25:702-11. [Crossref] [PubMed]
- Woo CG, Seo S, Kim SW, et al. Differential protein stability and clinical responses of EML4-ALK fusion variants to various ALK inhibitors in advanced ALK-rearranged non-small cell lung cancer. Ann Oncol 2017;28:791-7. [Crossref] [PubMed]
- Chang GC, Yang TY, Chen KC, et al. ALK variants, PD-L1 expression, and their association with outcomes in ALK-positive NSCLC patients. Sci Rep 2020;10:21063. [Crossref] [PubMed]
- Tao H, Shi L, Zhou A, et al. Distribution of EML4-ALK fusion variants and clinical outcomes in patients with resected non-small cell lung cancer. Lung Cancer 2020;149:154-61. [Crossref] [PubMed]
- Lin JJ, Zhu VW, Yoda S, et al. Impact of EML4-ALK Variant on Resistance Mechanisms and Clinical Outcomes in ALK-Positive Lung Cancer. J Clin Oncol 2018;36:1199-206. [Crossref] [PubMed]
- Christopoulos P, Kirchner M, Endris V, et al. EML4-ALK V3, treatment resistance, and survival: refining the diagnosis of ALK+ NSCLC. J Thorac Dis 2018;10:S1989-S1991. [Crossref] [PubMed]
- Yoda S, Lin JJ, Lawrence MS, et al. Sequential ALK Inhibitors Can Select for Lorlatinib-Resistant Compound ALK Mutations in ALK-Positive Lung Cancer. Cancer Discov 2018;8:714-29. [Crossref] [PubMed]
- Christopoulos P, Dietz S, Kirchner M, et al. Detection of TP53 Mutations in Tissue or Liquid Rebiopsies at Progression Identifies ALK+ Lung Cancer Patients with Poor Survival. Cancers (Basel) 2019;11:124. [Crossref] [PubMed]
- Christopoulos P, Budczies J, Kirchner M, et al. Defining molecular risk in ALK(+) NSCLC. Oncotarget 2019;10:3093-103. [Crossref] [PubMed]
- O'Regan L, Barone G, Adib R, et al. EML4-ALK V3 oncogenic fusion proteins promote microtubule stabilization and accelerated migration through NEK9 and NEK7. J Cell Sci 2020;133:jcs241505 [Crossref] [PubMed]
- Provencio-Pulla M, Serna R, Franco F, et al. ctDNA levels before treatment predict survival in non-small cell lung cancer patients treated with a tyrosine kinase inhibitor. J Clin Oncol 2020;38:9542. [Crossref]
- Kwon M, Ku BM, Park S, et al. Longitudinal monitoring by next generation sequencing of plasma cell-free DNA in ALK-rearranged non-small cell lung cancer (NSCLC) patients treated with ALK tyrosine kinase inhibitors. J Clin Oncol 2020;38:9603. [Crossref]
- Gray JE, Okamoto I, Sriuranpong V, et al. Tissue and Plasma EGFR Mutation Analysis in the FLAURA Trial: Osimertinib versus Comparator EGFR Tyrosine Kinase Inhibitor as First-Line Treatment in Patients with EGFR-Mutated Advanced Non-Small Cell Lung Cancer. Clin Cancer Res 2019;25:6644-52. [Crossref] [PubMed]
- Madsen AT, Winther-Larsen A, McCulloch T, et al. Genomic Profiling of Circulating Tumor DNA Predicts Outcome and Demonstrates Tumor Evolution in ALK-Positive Non-Small Cell Lung Cancer Patients. Cancers (Basel) 2020;12:947. [Crossref] [PubMed]
- Zhou C, Imamura F, Cheng Y, et al. Early clearance of plasma EGFR mutations as a predictor of response to osimertinib and comparator EGFR-TKIs in the FLAURA trial. J Clin Oncol 2019;37:9020. [Crossref]
- Shaw AT, Martini JF, Besse B, et al. Early circulating tumor (ct)DNA dynamics and efficacy of lorlatinib in patients (pts) with advanced ALK-positive non-small cell lung cancer (NSCLC). J Clin Oncol 2019;37:9019. [Crossref]
- Zhang LL, Kan M, Zhang MM, et al. Multiregion sequencing reveals the intratumor heterogeneity of driver mutations in TP53-driven non-small cell lung cancer. Int J Cancer 2017;140:103-8. [Crossref] [PubMed]
- Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med 2017;376:2109-21. [Crossref] [PubMed]
- Wu Z, Yang Z, Dai Y, et al. Update on liquid biopsy in clinical management of non-small cell lung cancer. Onco Targets Ther 2019;12:5097-109. [Crossref] [PubMed]
- Aldea M, Hendriks L, Mezquita L, et al. Circulating Tumor DNA Analysis for Patients with Oncogene-Addicted NSCLC With Isolated Central Nervous System Progression. J Thorac Oncol 2020;15:383-91. [Crossref] [PubMed]
- Boire A, Brandsma D, Brastianos PK, et al. Liquid biopsy in central nervous system metastases: a RANO review and proposals for clinical applications. Neuro Oncol 2019;21:571-84. [Crossref] [PubMed]
- Suryavanshi M, Jaipuria J, Panigrahi MK, et al. CSF cell-free DNA EGFR testing using DdPCR holds promise over conventional modalities for diagnosing leptomeningeal involvement in patients with non-small cell lung cancer. Lung Cancer 2020;148:33-9. [Crossref] [PubMed]
- Zheng MM, Li YS, Jiang BY, et al. Clinical Utility of Cerebrospinal Fluid Cell-Free DNA as Liquid Biopsy for Leptomeningeal Metastases in ALK-Rearranged NSCLC. J Thorac Oncol 2019;14:924-32. [Crossref] [PubMed]
- De Mattos-Arruda L, Mayor R, Ng CKY, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun 2015;6:8839. [Crossref] [PubMed]
- Shaw AT, Bauer TM, Marinis FD, et al. First-Line Lorlatinib or Crizotinib in Advanced ALK-Positive Lung Cancer. N Engl J Med 2020;383:2018-29. [Crossref] [PubMed]



