
Activity and mechanism of acquired resistance to tarloxotinib in HER2 mutant lung cancer: an in vitro study
Introduction
HER2 activating mutations, most of which are exon 20 insertion mutations, are present in 2–3% of lung adenocarcinomas (1,2). Despite the rapid development of molecular targeted therapies for non-small cell lung cancers (NSCLCs) with EGFR, ALK, ROS1, BRAF, NTRK or MET alterations, no drug has been approved for the treatment of HER2-mutant NSCLCs. Several clinical trials of HER2-targeted drugs, such as afatinib, neratinib and dacomitinib, for HER2-mutant NSCLC have been conducted; however, the activity of these drugs was limited (3-5).
Recently, we reported potent activity of poziotinib for HER2 exon 20 insertions by comparing nine HER2 inhibitors in an in vitro study (6). Poziotinib has shown potent activity against EGFR and HER2 exon 20 mutant lung cancer in phase II trials and preclinical studies (7,8); however, 67% (8/12) of patients who had received poziotinib experienced dose reduction due to adverse events, and eight patients (67%) had grade 3–4 AEs (8). Therefore, for HER2-mutated lung cancers, a variety of new therapeutic strategies are currently being tested in clinical trials, including a new drug delivery system (tarloxotinib) and an antibody-drug conjugates [trastuzumab deruxtecan (T-DXd)] (9).
Tarloxotinib is a novel pan-HER-targeted drug (10), and a phase II study of NSCLCs harboring HER2-activated mutations or EGFR exon 20 mutations is ongoing (NCT03805841). This drug is designed as a prodrug (tarloxotinib) that releases the activated pan-HER inhibitor [tarloxotinib-effector (tarloxotinib-E)] under pathological hypoxic conditions within the tumor (Figure S1) (11). This activation mechanism is expected to enlarge the therapeutic window and reduce toxicities such as diarrhea, rash and paronychia as a systemically active pan-HER inhibitor.
In this study, we evaluated the activity of tarloxotinib-E using in vitro models of HER2-mutated lung adenocarcinoma. In addition, we also investigated potential acquired resistance mechanisms to tarloxotinib-E. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/tlcr-21-216).
Methods
Cell lines
We used the immortalized murine pro-B cell line Ba/F3, which was obtained from the RIKEN Bio Resource Center (Tsukuba, Japan). NCI-H1781 (RRID:CVCL_1494) cells, which are a lung adenocarcinoma cell line harboring the HER2 exon 20 G776delins VC mutation (VC), were purchased from the American Type Culture Collection (Virginia, USA) and authenticated by short-tandem repeat profiling (Takara, Kusatsu, Japan). These cells were cultured in RPMI 1640 (Wako, Osaka, Japan) with 10% FBS (Sigma-Aldrich, St. Louis, MO, USA) and penicillin-streptomycin (P/S, Wako) at 37 °C with 5% CO2. We routinely checked for mycoplasma contamination using the TaKaRa PCR Mycoplasma Detection Set (Takara).
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All the in vitro experiments were performed in compliance with the institutional and national guidelines.
Establishment of Ba/F3 cells with HER2 mutations
Ba/F3 cells harboring the A775_G776insYVMA (YVMA), VC, or P780_Y781insGSP (GSP) HER2 mutation and Ba/F3 cells with wild-type (WT) HER2 were generated in our previous study (6,12). Ba/F3 cells with one of four HER2 point mutations (S310F, V659E, L755A, or L755P) were established using retroviral transfection as described previously (6,12). The list of designed primers is summarized in Table S1. Briefly, a pBABE retrovirus vector subcloned cDNA of the human WT HER2 gene was purchased from Addgene (Cambridge, MA, USA). We introduced each HER2 mutation into the pBABE vector of WT HER2 using a Prime STAR Mutagenesis Basal Kit (Takara) and designed primers (Table S1). The pBABE constructs were transfected with a pVSV-G vector [Takara (Clontech, Fremont, CA, USA), RRID:Addgene_138479] into gp-IRES 293 cells using FuGENE6 transfection reagent (Promega, Madison, WI, USA). Forty-eight hours after transfection, we collected the culture medium, and the viral particles were concentrated using a Retrovirus Concentration kit [Takara (Clontech)] and stored at −80 °C. Ba/F3 cells (3×103/well) were seeded onto 24-well plates and cultured for 24 hours. A retroviral suspension was added to each well. After a few days of incubation, the infected Ba/F3 cells were selected using puromycin (1.0–1.5 μg/mL) in the presence of interleukin-3 (IL-3). Then, we cultured puromycin-selected cells without IL-3 for approximately one week.
Reagents
Tarloxotinib and its activated drug, tarloxotinib-E, were provided by Rain Therapeutics Inc. We also used afatinib, poziotinib [second-generation (2G) EGFR TKIs], osimertinib [third-generation (3G) EGFR TKIs], and pyrotinib, which were purchased from Selleck Chemicals (Houston, TX, USA). Each drug was dissolved to a concentration of 10 mmol/L in dimethyl sulfoxide (DMSO, Sigma-Aldrich) and stored at −80 °C until use.
Cell proliferation assay
We seeded 3×104 transfected Ba/F3 cells into six-well plates. As a control, nontransfected Ba/F3 cells were also cultured in the presence and absence of IL-3. We counted the number of cells in each well using OneCell Counter (Biomedical Medical Science, Tokyo, Japan) in triplicate every 24 hours for 96 hours.
Cell viability assay
Cell viability assays were performed using Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan). A total of 3×103 cells were plated onto 96-well plates in technical triplicates and incubated for 24 hours. The cells were treated with each TKI at ten different drug concentrations. After 72 hours, 10 μL of tetrazolium salt WST-8 was added to each well, and plates were incubated for 2–4 hours. To determine bioluminescence, the formazan dye generated from tetrazolium in the reduction reaction with NADH, which reflects cell viability, was measured by reading the absorbance at 450 nm using a multiplate reader (Tecan, Mannedorf, Switzerland). Cell viability assay was performed at least twice.
Statistical analysis
The growth inhibitory curves were generated using GraphPad Prism 9 (GraphPad Software, San Diego, CA). Calculation of the standard deviation and estimation of the IC50 values were also performed using GraphPad Prism 9.
Generation of tarloxotinib-E resistant clones
We established tarloxotinib-E-resistant (TR) clones using HER2-mutant Ba/F3 cells through the N-ethyl-N-nitrosourea (ENU, Sigma-Aldrich) mutagenesis technique, as previously reported (13). HER2 mutant Ba/F3 cells were exposed to 100 μg/mL ENU for 24 hours. After removal of ENU, 1×105 cells were plated onto 96-well plates. These cells were incubated with 200 nM tarloxotinib-E for 14-21 days (the medium was changed every a few days), and TR clones were selected.
In the chronic exposure assay, H1781 cells were cultured with an increasing concentration of tarloxotinib-E starting at 2 nM. Acquired resistance was defined by the 50-fold higher IC50 of the TR cells than that of the parental H1781 cells.
HER2 mutation analyses
Total RNA from the transfected cells and resistant clones was extracted using a mirVana miRNA Isolation Kit (Qiagen, Hilden, Germany) and transcribed to cDNA using ReverTra Ace (TOYOBO, Osaka, Japan). Each transduced region and HER2 tyrosine kinase domains (TKD, exons 18 to 24) were amplified using designed primers. Sanger sequencing was conducted by a Genetic Analyzer 3130 or 3500XL (Applied Biosystems, Waltham, MA).
Western blot analysis
The cells were treated with the indicated concentrations of tarloxotinib-E or DMSO for 6 hours and washed with PBS. Total cell pellets were dissolved in lysis buffer, and a total of 15–30 μg of protein was loaded into SuperSep TM Ace (Wako) and blotted. Protein was transferred to the membrane [Trans-Blot® Turbo™ Mini PVDF Transfer Pack (Bio-Rad, Hercules, CA)], and membranes were probed using one of following antibodies: anti-HER2 (Cell Signaling Technology Cat# 2242, RRID:AB_331015), phosphorylated (p)-HER2 (Tyr1221/1222, Cell Signaling Technology Cat# 2243, RRID:AB_490899), anti-HER3 (Cell Signaling Technology Cat# 4754, RRID:AB_10691324), p-HER3 (Tyr1289, Cell Signaling Technology Cat# 4791, RRID:AB_2099709), anti-Akt (Cell Signaling Technology Cat# 9272, RRID:AB_329827), p-Akt (Ser473, Cell Signaling Technology Cat# 9271, RRID:AB_329825), anti-Erk1/2 (p44/42 MAPK, Cell Signaling Technology Cat# 9102, RRID:AB_330744), p-Erk1/2 (p44/42 MAPK, Thr202/Tyr204, Cell Signaling Technology Cat# 9100, RRID:AB_330741), and β-actin (Cell Signaling Technology Cat# 4967, RRID:AB_330288) overnight at 4 °C. Horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (Cell Signaling Technology Cat# 7074, RRID:AB_2099233) was used as the secondary antibody. All antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA). In the chemiluminescence assay, the membranes were reacted with enhanced chemiluminescence (ECL) solution (GE Healthcare, Chicago, Illinois) and scanned by an Amersham imager 680 (GE Healthcare).
p-receptor tyrosine kinase (RTK) array analysis
To detect the changes in the phosphorylation status of 49 different RTKs, we used a Human Phospho-RTK Array (R&D, Minneapolis, MS, USA). Parental H1781 and TR cells were treated with DMSO or 100 nM tarloxotinib-E for 24 hours, and then, cell lysates were prepared using lysis buffer according to the protocol. After blocking the array sheets, 200 µg of prepared sample was added to the array sheets and incubated overnight at 4 °C. After washing with wash buffer, the diluted anti-p-tyrosine-HRP detection antibody was pipetted into each array sheet. The array sheets were exposed to Chemi-Reagent Mix and scanned by an Amersham imager 680 (GE Healthcare).
Small interfering RNA
TR clones generated from H1781 cells were reverse-transfected with scrambled or HER3-targeting small interfering RNA (siRNA) purchased from Sigma-Aldrich (scramble siRNA, siHER3). Briefly, siRNA dissolved in OptiMEN (Thermo Fisher, Waltham, MA, USA) was spread to 60 mm dishes or 96-well plates, and H1781 TR cells were seeded in the plates. After 24 hours of incubation, H1781 TR cells were treated with tarloxotinib-E or DMSO. We collected H1781 TR cells after 12 hours of treatment and used them for western blotting.
Copy number analysis
Genomic DNA was extracted using a DNA extraction kit (Qiagen). Quantitative real-time PCR (qPCR) was performed using CYBR Green Real-time PCR Master Mix (Applied Biosystems) and the designed primers described in Table S2 with the StepOnePlus System (Applied Biosystems, Foster City, CA). The copy number of the LINE-1 gene was utilized as the internal control as described previously (14).
mRNA expression analysis
We extracted the total mRNA from H1781 and TR cells and then reverse-transcribed it to cDNA as described above. qPCR was performed using TaqMan probes targeting HER3 and TaqMan Fast Advanced Master Mix (Applied Biosystems). We quantitated the mRNA expression of HER3 by the delta-delta CT method using the expression of 18S ribosomal RNA as the internal control as described previously (14).
Results
HER2 mutations in non-small cell lung cancer
We searched for the frequency of HER2 mutations in NSCLC using the cBioPortal database (https://www.cbioportal.org) on March 1st, 2019 (Figure 1). To avoid overlapping data, we referred to eight studies to identify the common HER2 mutation in NSCLC (2,15-21). As previously reported (6,12), HER2 exon 20 insertions, YVMA, VC and GSP, were the most common HER2 mutations in NSCLC. In addition to these insertions, four single base substitutions, namely, S310F, V659E, L755A and L755P, were identified as common HER2 gene alternations in NSCLC.

In our previous study (6), we observed that Ba/F3 cells harboring YVMA, VC, GSP and WT HER2 proliferated well independent of IL-3. In this study, we found that Ba/F3 cells harboring one of the three point mutations (V659E, L755A and L755P) were also viable in the absence of IL-3 but not Ba/F3 cells with the S310F mutation (Figure S2).
Activity of tarloxotinib-E in HER2 mutated in vitro models
In the growth inhibitory assay, tarloxotinib-E showed potent efficacy against all Ba/F3 cells harboring HER2 exon 20 insertions as well as those harboring HER2 point mutations (Figure 2A,B). The IC50 values of tarloxotinib-E for HER2-mutant Ba/F3 cells and H1781 cells were less than 5 nM, which were lower than afatinib, osimertinib and pyrotinib and equivalent to that of poziotinib. The IC50 values of tarloxotinib (prodrug) for WT HER2 were much higher than those of other TKIs; for example, tarloxotinib had an 800-fold higher IC50 value than poziotinib. Western blotting showed that tarloxotinib-E inhibited the phosphorylation of HER2 exon 20 insertions and point mutations at 10 nM (Figure S3). On the other hand, tarloxotinib had no effect on the phosphorylation of HER2, even at a concentration of 100 nM (Figure S3).
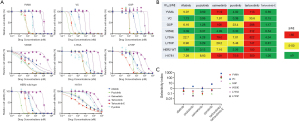
To compare the therapeutic window, we calculated the selectivity index (SI) of these drugs according to a previous report (22). The SI was calculated as the ratio of the IC50 of each TKI for WT HER2 and for each HER2 mutation (Figure 2C). Because tarloxotinib circulates as a prodrug and is activated under hypoxic intratumor conditions, the SI was indicated as [IC50 of tarloxotinib for WT HER2 cells]/[IC50 of tarloxotinib-E for HER2 mutant cells]. Many of the SIs of afatinib, poziotinib, osimertinib and pyrotinib were <1, meaning that these TKIs have inhibitory activity against WT HER2 compared with HER2 mutants. In contrast, the SI of tarloxotinib/tarloxotinib-E was >10 for all HER2-mutant Ba/F3 cells, indicating that tarloxotinib will have a wide therapeutic window in the clinical setting.
C805S mutation as an acquired resistance mechanism to tarloxotinib-E
To investigate the mechanisms of acquired resistance to tarloxotinib-E, we performed ENU mutagenesis using Ba/F3 cells with HER2 mutations. In this assay, we obtained a total of 50 clones that acquired tarloxotinib-E resistance from HER2-mutant Ba/F3 cells (Figure 3A). Thirty clones among these 50 developed HER2 secondary mutations, all of which were C805S substitutions, which are homologous to C797S of EGFR (Figure 3B). No secondary mutation was found in the HER2 TKD of the remaining 20 clones.
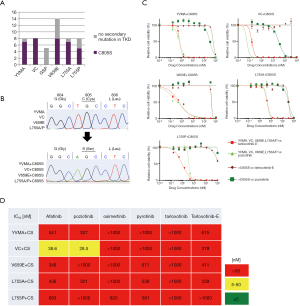
The IC50 values of tarloxotinib-E and poziotinib for Ba/F3 cells that acquired C805S were 50–200 times higher than those of parental cells (Figure 3C,3D). We also observed that Ba/F3 cells with secondary C805S mutations were resistant to all currently available HER2-TKIs (Figure 3D and Figure S4A). As shown in the immunoblots, the phosphorylation of HER2 in resistant cells treated with C805S was not reduced by tarloxotinib-E, even at a concentration of 100 nM (Figure S4B).
Activation of HER3 as an acquired resistance mechanism to tarloxotinib-E
We also established H1781 cells with acquired resistance to tarloxotinib-E (H1781 TR cells) via a chronic exposure assay at increasing concentrations of tarloxotinib-E from 2 nM to 100 nM. This clone showed approximately 100 times higher IC50 values than the parental H1781 cells (Figure 4A). There was no morphologic change that suggests the acquisition of epithelial to mesenchymal transition in H1781 TR cells (Figure 4B). HER2 phosphorylation was suppressed in H1781 TR cells, while Erk1/2 and Akt phosphorylation remained after 100 nM tarloxotinib-E treatment (Figure 4C). We detected no secondary mutations in the HER2 TKD, including C805S, in H1781 TR cells.
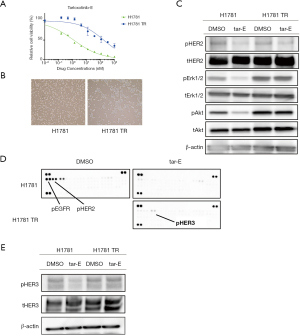
To explore the resistance mechanism of H1781 TR cells, we used the Human Phospho-RTK Array (Figure 4D). In this immunoassay, the phosphorylation of HER3 was increased in H1781 TR cells treated with 100 nM tarloxotinib-E compared with H1781 parental cells treated with 100 nM tarloxotinib-E. Using western blotting, we confirmed that p-HER3 was higher in H1781 TR cells than in parental cells in the presence of tarloxotinib-E (Figure 4E). We identified that the mRNA expression of HER3 was increased in TR cells compared with parental H1781 cells (Figure 5A). In addition, we found that H1781 TR cells acquired a gene copy number gain of HER3 by 1.5 times compared with parental cells (Figure 5B).
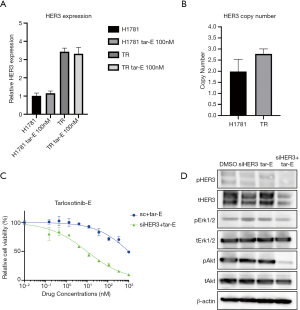
To analyze the role of HER3 in H1781 TR cells, we silenced HER3 using siRNA in this cell line. We observed that the combination of tarloxotinib-E and siRNA-mediated HER3 knockdown effectively killed H1781 TR cells (Figure 5C). Western blotting revealed that pAkt was significantly suppressed by the combination of tarloxotinib-E and siRNA-mediated HER3 knockdown (Figure 5D).
Discussion
In this in vitro study, tarloxotinib-E demonstrated potent activity against Ba/F3 models with HER2 exon 20 insertions or with HER2 point mutations that have been detected in NSCLCs. Recently, some novel agents have shown promising activity against HER2-altered NSCLCs in early-phase clinical trials. Socinski et al. (23) reported a 27.8% overall response rate (ORR) and 5.6-month progression-free survival (PFS) in twelve patients in a phase II study of poziotinib. T-DXd, a HER2-targeted antibody-drug conjugate, achieved a 61.9% ORR in eleven NSCLC patients with HER2 mutations in a dose-escalation phase I study (9). Most recently, pyrotinib showed efficacy against HER2-mutant lung adenocarcinomas with an ORR of 30.0% and a PFS of 6.9 months in a phase II study (24). However, it is also true that these novel HER2 inhibitors have high toxicity, such as diarrhea, skin rash, and drug-induced interstitial lung disease. In the early-phase trials mentioned above, dose reduction was needed in 67% of patients who received poziotinib and in 24% of patients who received T-DXd due to adverse events (8,25).
In this study, we demonstrated that tarloxotinib (prodrug) was not toxic against Ba/F3 cells with WT HER2 in contrast to other HER2 TKIs (afatinib, poziotinib, pyrotinib, and osimertinib). Because tarloxotinib-E (active form) inhibited the growth of Ba/F3 cells with HER2 mutations at a similar range as poziotinib, we expect high efficacy and lower toxicity of tarloxotinib in patients with HER2 mutations. In fact, in the first report of an ongoing phase II study of tarloxotinib in NSCLC patients, tumor response by RECIST was observed in 44% (4/9) of a cohort of NSCLC patients with a HER2-activating mutation, while dose reduction was required only in 21.7% (5/23) of patients in a whole cohort (NSCLCs harboring EGFR exon 20 insertion or HER2 mutations and solid tumors with NRG1/ERBB gene fusions) (26).
In this study, we also explored acquired resistance mechanisms to tarloxotinib-E in vitro. In the Ba/F3 models, we identified a secondary HER2 C805S mutation. HER2 C805 is homologous to C797 in the EGFR gene, in which irreversible TKIs form covalent bonds; therefore, it is reasonable that the C805S secondary mutation conferred resistance to all irreversible HER2 inhibitors, including tarloxotinib-E.
In addition to the secondary C805S mutation, we identified an acquired resistance mechanism by increased HER3 expression in H1781 cells. To the best of our knowledge, this is the first report of HER3 activation as an acquired resistance mechanism to HER2 inhibitors in lung cancer cells with HER2 mutations, although HER3 has been reportedly involved in the acquisition of resistance to gefitinib [an EGFR-TKI (27)] and alectinib [an ALK-TKI (24)]. Western blot analysis revealed an increased pAkt level in H1781 TR cells with HER3 activation (Figure 4C,4E), which is reasonable because HER3 contains six Tyr-Xaa-Xaa-Met (YXXM) consensus binding sites for the SH2 domains of the p85 regulatory subunit of PI3K (28). We found that siRNA-mediated HER3 knockdown resensitized H1781 TR cells to tarloxotinib-E, confirming the role of HER3 in the acquisition of resistance to tarloxotinib-E in H1781 cells. Therapeutically, we expect that anti-HER3 monoclonal antibodies, such as patritumab or lumretuzumab, or bispecific HER2/HER3 antibodies, such as istiratumab or duligotumab, may be able to overcome tarloxotinib-E resistance with HER3 activation (29). In this study, we explored acquired resistance mechanisms mediated by secondary HER2 mutations using Ba/F3 models and those mediated by bypass signaling using H1781 cells. However, it is not clear which mechanism, secondary mutations vs. bypass pathway, is more likely to occur in the actual treatment of HER2-mutated lung cancer patients. In addition, it is possible that other types of resistances, such as histological transformation or modification of tumor microenvironment, may arise. Therefore, future analyses using clinical specimens obtained from tarloxotinib-E refractory patients are necessary to confirm our in vitro findings.
In conclusion, tarloxotinib-E showed potent activity against cell lines expressing HER2 mutations, both exon 20 insertions and HER2 point mutations. In addition, we identified a secondary C805S HER2 mutation as well as increased HER3 expression as molecular mechanisms of acquired resistance to tarloxotinib-E in our in vitro models.
Acknowledgments
The authors thank Ms. Keiko Obata (Department of Surgery, Kindai University Faculty of Medicine) for her technical assistance. We thank American Journal Express (
Funding: This study was supported by grants-in-aid for scientific research from the Japan Society for Promotion of Science (grant 19K16785 to Dr. Koga, grant 18K07336 to Dr. Suda, grant 19K09285 to Dr. Soh) and a research grant from Rain Therapeutics to Dr. Suda.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/tlcr-21-216
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tlcr-21-216
Peer Review File: Available at https://dx.doi.org/10.21037/tlcr-21-216
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tlcr-21-216). Dr. TM serves as an unpaid editorial board member of Translational Lung Cancer Research from Sep 2019 to Sep 2021. Dr. TK has received research funding from Boehringer Ingelheim outside the submitted work. Dr. KS has received research funding from Rain Therapeutics and Boehringer Ingelheim, an honorarium from Boehringer Ingelheim, AstraZeneca, and Chugai outside the submitted work, and has been on the advisory board of AstraZeneca. Dr. TF has received research funding from Apollomics and an honorarium form Novartis outside the submitted work. Dr. AH has received lecture fees from AstraZeneca and Chugai outside the submitted work. Dr. VT and Mr. AV are currently employees of rain therapeutics and own stock in rain therapeutics. Dr. RCD is currently an employee of Rain Therapeutics and owns stock in Rain Therapeutics and has received personal fees from Genentech/Roche, Ignyta, Blueprint Medicines, Green Peptide, AstraZeneca, Anchiano, Takeda/Millenium, and Bayer outside the submitted work; in addition, Dr. RCD has a patent for U.S. Provisional Patent Application No. 62/712,531 pending and licensed to Rain Therapeutics. The University of Colorado has received licensing fees from Foundation Medicine, Ignyta, Scorpion Therapeutics, Voronoi, Pearl River, Black Diamond Therapeutics, and Genentech for biologic materials derived in Dr. RCD’s laboratory. Dr. TM has received research funding from Astra Zeneca, Boehringer Ingelheim, Chugai, Taiho, Ono Pharmaceutical, Merck Sharp and Dohme, and Eli-Lilly; has been on the advisory board of Novartis, AstraZeneca, Amgen, Janssen pharma, Merck Sharp and Dohme and Puma Biotech; has received lecture fees from AstraZeneca, Boehringer Ingelheim, Novartis, Chugai, Pfizer, Bristol-Myers Squibb, Eli Lilly, Merck Sharp and Dohme, Ono Pharmaceutical, Merck Biopharma and Takeda, outside the submitted work. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All the
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hirsch FR, Suda K, Wiens J, et al. New and emerging targeted treatments in advanced non-small-cell lung cancer. Lancet 2016;388:1012-24. [Crossref] [PubMed]
- Jordan EJ, Kim HR, Arcila ME, et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov 2017;7:596-609. [Crossref] [PubMed]
- Hyman DM, Piha-Paul SA, Won H, et al. HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature 2018;554:189-94. [Crossref] [PubMed]
- Kris MG, Camidge DR, Giaccone G, et al. Targeting HER2 aberrations as actionable drivers in lung cancers: phase II trial of the pan-HER tyrosine kinase inhibitor dacomitinib in patients with HER2-mutant or amplified tumors. Ann Oncol 2015;26:1421-7. [Crossref] [PubMed]
- Dziadziuszko R, Smit EF, Dafni U, et al. Afatinib in NSCLC With HER2 Mutations: Results of the Prospective, Open-Label Phase II NICHE Trial of European Thoracic Oncology Platform (ETOP). J Thorac Oncol 2019;14:1086-94. [Crossref] [PubMed]
- Koga T, Kobayashi Y, Tomizawa K, et al. Activity of a novel HER2 inhibitor, poziotinib, for HER2 exon 20 mutations in lung cancer and mechanism of acquired resistance: An in vitro study. Lung Cancer 2018;126:72-9. [Crossref] [PubMed]
- Robichaux JP, Elamin YY, Tan Z, et al. Mechanisms and clinical activity of an EGFR and HER2 exon 20-selective kinase inhibitor in non-small cell lung cancer. Nat Med 2018;24:638-46. [Crossref] [PubMed]
- Robichaux JP, Elamin YY, Vijayan RSK, et al. Pan-Cancer Landscape and Analysis of ERBB2 Mutations Identifies Poziotinib as a Clinically Active Inhibitor and Enhancer of T-DM1 Activity. Cancer Cell 2019;36:444-457.e7. [Crossref] [PubMed]
- Smit EF, Nakagawa K, Nagasaka M, et al. Trastuzumab deruxtecan (T-DXd; DS-8201) in patients with HER2-mutated metastatic non-small cell lung cancer (NSCLC): Interim results of DESTINY-Lung01. J Clin Oncol 2020;38:9504. [Crossref]
- Estrada-Bernal A, Le AT, Doak AE, et al. Tarloxotinib Is a Hypoxia-Activated Pan-HER Kinase Inhibitor Active Against a Broad Range of HER-Family Oncogenes. Clin Cancer Res 2021;27:1463-75. [Crossref] [PubMed]
- Jing X, Yang F, Shao C, et al. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol Cancer 2019;18:157. [Crossref] [PubMed]
- Kosaka T, Tanizaki J, Paranal RM, et al. Response Heterogeneity of EGFR and HER2 Exon 20 Insertions to Covalent EGFR and HER2 Inhibitors. Cancer Res 2017;77:2712-21. [Crossref] [PubMed]
- Kobayashi Y, Azuma K, Nagai H, et al. Characterization of EGFR T790M, L792F, and C797S Mutations as Mechanisms of Acquired Resistance to Afatinib in Lung Cancer. Mol Cancer Ther 2017;16:357-64. [Crossref] [PubMed]
- Suda K, Murakami I, Katayama T, et al. Reciprocal and complementary role of MET amplification and EGFR T790M mutation in acquired resistance to kinase inhibitors in lung cancer. Clin Cancer Res 2010;16:5489-98. [Crossref] [PubMed]
- Rizvi H, Sanchez-Vega F, La K, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol 2018;36:633-41. [Crossref] [PubMed]
- Vavalà T, Monica V, Lo Iacono M, et al. Precision medicine in age-specific non-small-cell-lung-cancer patients: Integrating biomolecular results into clinical practice-A new approach to improve personalized translational research. Lung Cancer 2017;107:84-90. [Crossref] [PubMed]
- Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607-16. [Crossref] [PubMed]
- Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012;150:1107-20. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069-75. [Crossref] [PubMed]
- Udagawa H, Hasako S, Ohashi A, et al. TAS6417/CLN-081 Is a Pan-Mutation-Selective EGFR Tyrosine Kinase Inhibitor with a Broad Spectrum of Preclinical Activity against Clinically Relevant EGFR Mutations. Mol Cancer Res 2019;17:2233-43. [Crossref] [PubMed]
- Socinski MA, Cornelissen R, Garassino MC, et al. LBA60 ZENITH20, a multinational, multi-cohort phase II study of poziotinib in NSCLC patients with EGFR or HER2 exon 20 insertion mutations. Ann Oncol 2020;31:S1188. [Crossref]
- Zhou C, Li X, Wang Q, et al. Pyrotinib in HER2-Mutant Advanced Lung Adenocarcinoma After Platinum-Based Chemotherapy: A Multicenter, Open-Label, Single-Arm, Phase II Study. J Clin Oncol 2020;38:2753-61. [Crossref] [PubMed]
- Tsurutani J, Iwata H, Krop I, et al. Targeting HER2 with Trastuzumab Deruxtecan: A Dose-Expansion, Phase I Study in Multiple Advanced Solid Tumors. Cancer Discov 2020;10:688-701. [Crossref] [PubMed]
- Liu SV, Villaruz LC, Lee VHF, et al. LBA61 first analysis of RAIN-701: study of tarloxotinib in patients with non-small cell lung cancer (NSCLC) EGFR Exon 20 insertion, HER2-activating mutations & other solid tumours with NRG1/ERBB gene fusions. Ann Oncol 2020;31:S1189. [Crossref]
- Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039-43. [Crossref] [PubMed]
- Hsieh AC, Moasser MM. Targeting HER proteins in cancer therapy and the role of the non-target HER3. Br J Cancer 2007;97:453-7. [Crossref] [PubMed]
- Liu X, Liu S, Lyu H, et al. Development of Effective Therapeutics Targeting HER3 for Cancer Treatment. Biol Proced Online 2019;21:5. [Crossref] [PubMed]


