
A large-scale, multicentered trial evaluating the sensitivity and specificity of digital PCR versus ARMS-PCR for detecting ctDNA-based EGFR p.T790M in non-small-cell lung cancer patients
Introduction
The identification of epidermal growth factor receptor (EGFR) mutations has led to dramatic changes in the treatment of a variety of cancers, especially non-small cell lung cancer (NSCLC) (1-3). The exploration of EGFR tyrosine kinase inhibitors (TKIs) has brought significant improvement in progression-free survival (PFS), overall survival (OS), and quality of life compared to standard chemotherapy to patients with EGFR-mutated NSCLC (4-10). Despite the significant initial benefit of the first and second-generation EGFR-TKIs, resistance commonly develops within 10–16 months, with the occurrence of EGFR p.T790M accounting for 50–65% of the resistance to EGFR-TKIs (11-15). Osimertinib, the third-generation TKI developed to overcome the EGFR p.T790M-induced resistance, is commonly administrated to NSCLC patients with EGFR-activating mutations in the first-line setting or patients with the EGFR p.T790M after progression on first-line treatment of other EGFR-TKIs (16-21). Therefore, the detection and monitoring of EGFR p.T790M during diagnosis or treatment is vitally important for better and timely management of EGFR-mutated NSCLC patients.
Currently, the circulating tumor DNA (ctDNA) in peripheral blood has been established as an important surrogate of tumor tissues due to its mutation detection capabilities and less-invasive collection. Previous studies have proved the feasibility of using ctDNA-based EGFR mutation tests to guide first-line EGFR-TKI therapy and demonstrated the predictive potential of longitudinal monitoring of ctDNA during treatment (22,23). However, it is noted that ctDNA tests are highly specific yet not sensitive enough for companion diagnostic use. Thus, NCCN guidelines suggest that plasma-based testing should be considered at progression on EGFR-TKIs for p.T790M. If plasma-based testing is negative, tissue-based testing under re-biopsy is strongly recommended. Therefore, it is crucial to develop detection methods with higher sensitivity while maintaining specificity.
Several methods have been developed to routinely assess the EGFR mutations, such as the PCR-based technologies and next-generation sequencing (NGS) (24). The PCR-based technologies for detecting EGFR mutations contain digital PCR (dPCR) and amplification refractory mutation system (ARMS)-PCR, including ADx-ARMS® EGFR mutation detection kit of Amoy Diagnostics with a limit of detection (LoD) of 1%, and Roche Diagnostics’ Cobas-ARMS PCR kit (Cobas® EGFR Mutation Test v2) with an LoD of 0.2%, both according to the manufacturers’ protocols. dPCR technology was developed with the goal of improving the sensitivity of ARMS-PCR without compromising the specificity, which have been demonstrated by increasing publications (24-29). We have established the LoD and baseline for the EGFR p.T790M dPCR assay to be 0.1% with at least 5 mutant positive signals (30). Additionally, dPCR can perform absolute quantification without making a standard curve of an internal standard, and it does not depend on the PCR cyclic threshold (Ct) value for quantification. Hence, the superior sensitivity, high accuracy, and specificity makes dPCR especially suitable for measuring small amounts of ctDNA and extremely low allele frequency mutations in plasma samples. These measures of accuracy would make dPCR a strong candidate for liquid biopsy tests for companion diagnostics.
In order to demonstrate the sensitivity and specificity of dPCR assays for EGFR p.T790M detection, we initiated this clinical trial enrolling over 1,000 mNSCLC patients resistant to 1st-generation TKIs. In this study, we used chip-based dPCR technology to determine EGFR p.T790M status in plasma samples and compared the results to ADx-ARMS EGFR mutation detection kit. The Roche Diagnostics’ Cobas-ARMS kit (Cobas® EGFR Mutation Test v2) was used as a third method when digital PCR results disagree with comparator data. Paired tissue and plasma samples were also used to evaluate the accuracy of dPCR results. Lastly, we followed up on the clinical responses of osimertinib in EGFR p.T790M-positive patients as detected by either dPCR or ADx-ARMS PCR. It is hypothesized that dPCR will detect more positive samples than ADx-ARMS PCR and therefore would have better sensitivity than ADx-ARMS PCR. We present the following article in accordance with STARD reporting checklist (available at https://dx.doi.org/10.21037/tlcr-21-564).
Methods
Study design and participants
This prospective study (Chinese Clinical Trial Registry identifier: ChiCTR2100043147) included 1045 patients enrolled from 15 medical centers across China, including 1029 patients diagnosed with advanced NSCLC and 16 patients with specific samples (Table 1). Of the specific samples, 4 patients had lung nodules and 12 patients had small-cell-lung-cancer. All patients showed resistance to first-generation TKIs and had not received third generation TKIs. Collection was from April 1st, 2018 to November 30th, 2019. The plasma samples were collected once from all the patients enrolled, 45 of which also had paired tissue and plasma samples collected from the same patients at the same time period. The samples were tested for EGFR p.T790M by dPCR and ADx-ARMS® EGFR mutation detection kit (ADx-ARMS PCR) as a comparator assay; Cobas®EGFR Mutation Test v2 (Cobas PCR) was utilized for the samples with inconsistent results between ADx-ARMS PCR and dPCR.
Table 1
| Clinicopathological characteristics of patients | Number |
|---|---|
| Age | |
| Mean ± Std | 61.74±10.75 |
| Median (min–max) | 63.00 (24.00–91.00) |
| Age, years | |
| Under 50 | 137 (13.35%) |
| 50–59 | 268 (26.12%) |
| 60–69 | 392 (38.21%) |
| Above 69 | 229 (22.32%) |
| Gender | |
| Male | 438 (42.69%) |
| Female | 588 (57.31%) |
| Histology | |
| Adenocarcinoma | 806 (78.56%) |
| Squamous cell carcinoma | 11 (1.07%) |
| Adenocarcinoma mixed squamous cell carcinoma | 7 (0.68%) |
| Pulmonary nodules | 4 (0.39%) |
| Small cell lung cancer | 12 (1.17%) |
| Unknown | 186 (18.13%) |
| Tumor stage | |
| IA | 1 (0.10%) |
| IIB | 1 (0.10%) |
| III | 12 (1.17%) |
| IIIA | 21 (2.05%) |
| IIIB | 26 (2.53%) |
| IIIC | 2 (0.19%) |
| IV | 533 (51.95%) |
| IVA | 54 (5.26%) |
| IVB | 61 (5.95%) |
| Extensive period | 5 (0.49%) |
| Unknown | 310 (30.21%) |
| Resistant to EGFR-TKI | |
| Yes | 889 (86.65%) |
| No | 13 (1.27%) |
| Unknown | 124 (12.09%) |
Unknow: the outpatient doctors judged that the patients met the inclusion criteria of this clinical trial through the other hospital information shown by the patients, but we failed to find the relevant information of the patients from the Hospital Information System when tracing the information at last. EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor.
ADx-ARMS PCR was chosen as the primary reference assay as it was the only NMPA approved liquid biopsy assay at the time of the study. Sensitivity, specificity, PPV, NPV and overall concordance were analyzed against comparator assays. Additionally, efficacy was evaluated for patients with EGFR p.T790M detected by either dPCR or ADx-ARMS PCR, who underwent Osimertinib treatment, according to the RECIST 1.1 criteria.
This study was approved by the Institutional Review Board or Independent Ethics Committee at each institution and conducted in accordance with local regulatory requirements. The general principles of the International Ethical Guidelines for Biomedical Research Involving Human Subjects, the International Conference on Harmonisation guidelines on Good Clinical Practice, and the Declaration of Helsinki (as revised in 2013) were followed. All participants had signed informed consent. The full study protocol was reviewed by all hospital involved in this study and can be accessed via those hospitals.
Sample collection and DNA purification
For plasma samples, 20 mL peripheral blood samples were collected in two tubes using PAXgene Blood cfDNA Tube (catalog# 768115, QIAGEN, Hilden, Germany). The tubes were centrifuged at 1,600 g for 10 min to obtain plasma, and the plasma was centrifuged again at 1,600 g for 10 min to pellet debris. cfDNA was subsequently purified from the supernatant of the second centrifugation using the QIAamp Circulating Nucleic Acid Kit (catalog# 55114, QIAGEN). Each sample was eluted with 100 µL buffer AVE and concentrated to 20 µL using the DNA Clean & Concentrator-5 kit (catalog# D4013, Zymo Research, Irvine, CA, USA). Formalin fixed paraffin embedded (FFPE) tissues were obtained from 45 patients, and DNA was purified from the tissue using the QIAamp DNA FFPE Tissue Kit (catalog# 56404, QIAGEN). Purified double stranded genomic DNA was quantitated using the Qubit dsDNA HS Assay (catalog# Q32851, Thermo Fisher Scientific, Waltham, MA, USA) and Qubit™ 4 Fluorometer (catalog# Q33238, Thermo Fisher Scientific). The DNA was tested immediately or stored at −20 °C for later use.
EGFR p.T790M mutation detection
dPCR-based EGFR p.T790M detection kit (catalog# Q0242310, Questgenomics, Nanjing, China) was used to detect EGFR p.T790M mutation in plasma circulating tumor DNA (ctDNA). 14.5 µL reaction mixture was prepared consisting of 7.25 µL digital PCR reaction mix, 0.72 µL EGFR T790M mutation detection assay containing PCR primers and Taqman probes, 6.53 µL DNA sample containing 20 to 80 ng cfDNA. Two TaqMan probes with different fluorescent dyes were used to target wild-type (VIC-labeled) and T790M mutated EGFR sequences (FAM-labeled). The reaction mixture was loaded onto a silicon chip using a QuantStudio™ 3D Digital PCR Chip Loader (catalog# 4482592, Thermo Fisher Scientific), and the PCR reaction was performed on a ProFlex™ 2x Flat PCR System (catalog# 4484078, Thermo Fisher Scientific). Thermal cycling conditions were: 96 °C for 10 min; 60 °C for 2 min; 39 cycles of 98 °C for 30 seconds, 60 °C for 2 min; 60 °C for 2 min then 10 °C hold. Data were collected and processed using a QuantStudio™ 3D Digital PCR Instrument (catalog# 4489084, Thermo Fisher Scientific). The number of FAM-positive (EGFR p.T790M mutant allele) and VIC-positive (EGFR wild type allele) wells were counted in each sample. The number of target DNA copies was converted from the number of wells using the Poisson distribution. Mutant allele frequency was calculated as:
The cut-off value used was the LoD of EGFR p.T790M digital PCR assay at 0.1% mutant allele frequency (30). The receiver operating characteristic curve (ROC curve) was used to validate the cut-off value in this study.
The ADx-ARMS® EGFR mutation detection kit (catalog # ADx-EG01, Amoy Diagnostics, Xiamen, China) was used as a comparator assay and Cobas®EGFR Mutation Test v2 (catalog# 07248563190, Roche Molecular Systems, Inc., Pleasanton, CA) were used as a third method when digital PCR results are not in agreement with results from ADx-ARMS PCR. All assays used the same amount input cfDNA. The experimental conditions and data analysis followed the manufacturer’s instructions.
Statistical analysis
A blind analysis was conducted, and all statistical analyses were performed using SPSS Statistics (version 22). The agreement rate between the two test methods for detecting EGFR p.T790M was assessed by calculating the sensitivity, specificity, and concordance. The kappa value, an internal consistency coefficient, is used to evaluate concordance between the two methods. Kappa ≥0.75, 0.75> Kappa ≥0.4, and Kappa <0.4 indicate good, general, and poor consistency, respectively, between the two methods. Data were presented as mean ± standard deviation. A P value <0.05 was considered to be statistically significant.
Results
Patients and study framework
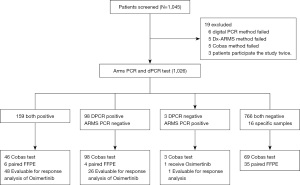
For this study, 1,045 patients were screened; 19 were excluded due to various assay test failures resulting in a grand total of 1,026 patients (Figure 1). Of the enrolled patients, the median age was 63.0 years old, 438 (42.69%) were males, 588 (57.31%) were females, and 709 (69.10%) had stage III–IV NSCLC (Table 1); 813 out of 1,026 patients (79.24%) were diagnosed as adenocarcinoma or had mixed squamous cell histology; 889 patients (86.69%) had evidence for progression on EGFR-TKI treatment. Plasma samples, and tissue samples if applicable, were taken at one point in time for all patients. None of the enrolled patients exhibited adverse events caused by the testing.
Comparison of EGFR p.T790M detection using dPCR, ADx-ARMS PCR, and Cobas PCR
In order to reduce incidences of false positive determination in liquid biopsy under the assumption that the sensitivity of liquid biopsy is not as good as tissue biopsy, the limit of detection at 0.1% mutant allele frequency was determined to be the cut-off value for EGFR p.T790M (30). At this cut-off value, the sensitivity, the specificity, the positive predictive value (PPV), the negative predictive value (NPV), and the concordance were calculated for the results of 1026 patients detected by dPCR assay and the NMPA-approved ADx-ARMS PCR, which was chosen as the reference method. As shown in Table 2, the sensitivity between the two methods was 98.15% (95% CI: 94.68–99.62%), the specificity was 88.66% (95% CI: 86.35–90.70%), the PPV was 61.87% (95% CI: 55.63–67.83%), the NPV was 99.61% (95% CI: 98.86–99.92%) and the concordance was 90.16% (95% CI: 88.17–91.91%). Among the 1026 samples, there are 4 patients with lung nodules and 12 patients with small-cell-lung-cancer enrolled as specific samples. As expected, all 16 samples were tested to be EGFR p.T790M negative.
Table 2
| Items | Cut-off value | N (%) | Discordant cases | Youden index | Sensitivity (95% CI) |
Specificity (95% CI) |
PPV (95% CI) | NPV (95% CI) | Concordance (95% CI) |
|---|---|---|---|---|---|---|---|---|---|
| dPCR vs. ADx-ARMS | 0.10% | 1,026 | 101 (9.84%) | 0.8681 | 98.15% (94.68–99.62%) |
88.66% (86.35–90.70%) |
61.87% (55.63–67.83%) |
99.61% (98.86–99.92%) |
90.16% (88.17–91.91%) |
| dPCR+/ADx-ARMS + | 159 (15.50%) | ||||||||
| dPCR-/ADx-ARMS + | 3 (0.29%) | ||||||||
| dPCR+/ADx-ARMS – | 98 (9.55%) | ||||||||
| dPCR-/ADx-ARMS – | 766 (74.66%) | ||||||||
| dPCR vs. ADx-ARMS | 0.11% | 1,026 | 95 (9.26%) |
0.8700 | 97.53% (93.80–99.32%) |
89.47% (87.23–91.44%) |
63.45% (57.14–69.44%) |
99.49% (98.69–99.86%) |
90.74% (88.80–92.44%) |
| dPCR+/ADx-ARMS + | 158 (15.40%) | ||||||||
| dPCR-/ADx-ARMS + | 4 (0.39%) | ||||||||
| dPCR+/ADx-ARMS – | 91 (8.87%) | ||||||||
| dPCR-/ADx-ARMS – | 773 (75.34%) |
EGFR, epidermal growth factor receptor; dPCR, digital polymerase chain reaction; ARMS-PCR, amplification refractory mutation system-polymerase chain reaction; CI, confidence interval; PPV, positive predictive value; NPV, the negative predictive value.
There was an additional 9.26% of patients detected positive by dPCR (257, 25.05%) compared to that by the ADx-ARMS PCR (162, 15.79%). The distribution of mutant allele frequency for dPCR positive, ADx-ARMS PCR negative, and dPCR positive, ADx-ARMS PCR negative samples are shown in Figure 2A. The distribution is much wider for dPCR positive, ADx-ARMS PCR positive samples with a range of 0.1–28.93% mutant allele frequency for the 159 patients. The distribution of dPCR positive, ADx-ARMS PCR negative samples is smaller in comparison, with all mutant allele frequencies below 10%. This shows that while both dPCR and ADx-ARMS PCR can detect high allele frequencies, dPCR is better at detecting low allele frequencies.
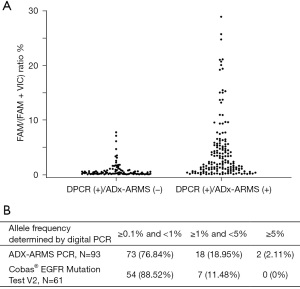
The Cobas®EGFR Mutation Test V2 from Roche, another qPCR-based comparator kit, was used as a third testing method on the 99 samples with non-concordant results between dPCR and the ADx-ARMS PCR. Among the 96 samples tested positive by dPCR, negative by ADx-ARMS PCR, 32 were tested positive by Cobas PCR. While the allele frequency cannot be determined using ADx-ARMS PCR or Cobas PCR, as they both used relative quantification, the allele frequencies determined by dPCR can be used to compare the performance of the two assays. For Cobas PCR samples, 88.52% of the non-concordant samples had a mutant allele frequency between 0.1% and 1% versus ADx-ARMS PCR which had only 76.84% of samples between that allele frequency range (Figure 2B). Looking further, Cobas PCR only had 11.48% of the non-concordant samples with an allele frequency between 1.0% and 5.0% while ADx-ARMS PCR had 18.95% of samples between that allele frequency range. Cobas PCR had no negative samples above 5.0% but ADx-ARMS PCR had 2.11%. These results show that Cobas PCR is more consistent at low allele frequencies than ADx-ARMS PCR.
An additional 115 were also analyzed by Cobas PCR to investigate the consistency between Cobas PCR and ADx-ARMS PCR when dPCR results are in agreement with ADx-ARMS PCR. Those samples included samples tested negative by both digital PCR and ADx-ARMS PCR, as well as samples tested positive by both methods that had low allele frequencies according to dPCR results. Test results by Cobas PCR are all concordant with previous results, suggesting Cobas PCR and ADx-ARMS PCR would have the same results if digital PCR and ADx-ARMS PCR has concordant results. Because these samples were concordant between all three assays, the other 812 sample results, out of the total 1,026 samples, were not reanalyzed by Cobas PCR because they were assumed to be concordant. The results of 115 concordant and 96 non-concordant samples analyzed by Cobas PCR replaced the results of those samples from ADx-ARMS PCR; this allows for the comparison of equivalent Cobas PCR results and dPCR results. dPCR found 6.82% more samples positive than qPCR; the sensitivity between the two methods was 98.40% (95% CI: 95.38–99.67%), the specificity was 91.30% (95% CI: 89.18–93.12%), PPV was 71.60% (95% CI: 65.66–77.03%), NPV was 99.61% (95% CI: 98.86–99.92%), and concordance was 92.59% (95% CI: 90.82–94.12%, Table 3). The inclusion of Cobas PCR resulted in improvement of the sensitivity and specificity to the results from ADx-ARMS PCR as shown in Table 2.
Table 3
| Items | Cut-off value | N (%) | Discordant cases | Youden index | Sensitivity (95% CI) |
Specificity (95% CI) |
PPV (95% CI) | NPV (95% CI) | Concordance (95% CI) |
|---|---|---|---|---|---|---|---|---|---|
| dPCR vs. ADx-ARMS +COBAS | 0.10% | 1,026 | 76 (7.41%) | 0.8970 | 98.40% (95.38–99.67%) |
91.30% (89.18–93.12%) |
71.60% (65.66–77.03%) |
99.61% (98.86–99.92%) |
92.59% (90.82–94.12%) |
| dPCR+/ADx-ARMS and cobas+ | 184 (17.93%) | ||||||||
| dPCR-/ADx-ARMS and cobas + | 3 (0.29%) | ||||||||
| dPCR+/ADx-ARMS and cobas- | 73 (7.12%) | ||||||||
| dPCR-/ADx-ARMS and cobas- | 766 (74.66%) |
EGFR, epidermal growth factor receptor; dPCR, digital polymerase chain reaction; ARMS-PCR, amplification refractory mutation system-polymerase chain reaction; CI, confidence interval; PPV, positive predictive value; NPV, the negative predictive value.
From these results it is clear that none of the tests agree on the number of positives; furthermore, Cobas PCR has better sensitivity than ADx-ARMS PCR not only because it was capable of detecting more positive, but more importantly, it was capable of detecting more positive samples at the allele frequency of above 1%. It became clear that at lower allele frequency (0.1% and 1%), both qPCR methods would have false negative results due to lack of sensitivity. Additionally, this implies that the disagreement between dPCR and ADx-ARMS PCR may not demonstrate that additional positive samples detected by dPCR are false negative samples.
Comparison of EGFR p.T790M detection in paired tissue and plasma samples using digital PCR and ARMS-PCR
Forty-five out of 1026 NSCLC patients enrolled had paired plasma and tissue samples. When compared to tissue biopsy results, the sensitivity, specificity, and concordance for the dPCR detection of EGFR p.T790M were 53.85% (95% CI: 25.13–80.78%), 90.63% (95% CI: 74.98–98.02%) and 80.00% (95% CI: 65.40–90.42%), respectively. When the EGFR p.T790M status of the 45 plasma samples with paired tissue samples was tested by ADx-ARMS PCR and compared to tissue biopsy results, the sensitivity, specificity, and concordance were 30.77% (95% CI: 9.09–61.43%), 93.75% (95% CI: 79.19–99.23%) and 75.56% (95% CI: 60.46–87.12%), respectively (Table 4). For Cobas PCR results compared against tissue biopsy results, 3 non-concordant samples were tested, and all were determined negative, suggested in this sample pool, the sensitivity of Cobas PCR is same as ADx-ARMS PCR at 30.77% (95% CI: 9.09%). The paired tissue and plasma samples demonstrate that dPCR is more sensitive than ADx-ARMS PCR with improved overall concordance. Specificity is still maintained at a level greater than 90%.
Table 4
| Items | Cut-off value | N (%) | Discordant cases | Sensitivity (95% CI) |
Specificity (95% CI) |
PPV (95% CI) | NPV (95% CI) | Concordance (95% CI) |
|---|---|---|---|---|---|---|---|---|
| dPCR | 0.10% | 45 | 9 (20.00%) | 53.85% (25.13–80.78%) |
90.63% (74.98–98.02%) |
70.00% (34.75–93.33%) |
82.86% (66.35–93.44%) |
80.00% (65.40–90.42%) |
| ctDNA+/FFPE + | 7 (15.56%) | |||||||
| ctDNA+/FFPE- | 3 (6.67%) | |||||||
| ctDNA-/FFPE+ | 6 (13.33%) | |||||||
| ctDNA-/FFPE- | 29 (64.44%) | |||||||
| ADx-ARMS | N/A | 45 | 11 (24.44%) | 30.77% (9.09–61.43%) |
93.75% (79.19–99.23%) |
66.67% (22.28–95.67%) |
76.92% (60.67–88.87%) |
75.56% (60.46–87.12%) |
| ctDNA+/FFPE + | 4 (8.89%) | |||||||
| ctDNA+/FFPE- | 2 (4.44%) | |||||||
| ctDNA-/FFPE+ | 9 (20.00%) | |||||||
| ctDNA-/FFPE- | 30 (66.67%) |
EGFR, epidermal growth factor receptor; dPCR, digital polymerase chain reaction; ARMS-PCR, amplification refractory mutation system-polymerase chain reaction; CI, confidence interval; PPV, positive predictive value; NPV, the negative predictive value; ctDNA, circulating tumor DNA; FFPE, formalin fixed paraffin-embedded tissue.
Taking a closer look at the results, among the 13 out of 45 samples tested positive in tissue biopsy, only 4 were tested positive in plasma samples by ADx-ARMS PCR. 4 were tested positive by Cobas PCR, and 7 were tested positive by dPCR. The additional 3 positive samples determined negative by ADx-ARMS PCR and positive by dPCR had an allele frequency of 0.21%, 0.94% and 1.66%, which is at an allele frequency close to or below the limit of detection of ADx-ARMS PCR. This validates the notion that these positive samples are real; the additional positive samples are confirmed to be true positives by comparing their status to tissue biopsy.
As expected due to tumor heterogeneity, there were 2 plasma samples tested positive by ADx-ARMS PCR and additional 1 sample tested positive by dPCR while the 32 FFPE samples exhibited negative results. The additional 1 sample tested positive by digital PCR but negative by ADx-ARMS PCR had a mutant allele frequency of 0.29%. The other two samples that had concordant results from ADx-ARMS PCR and dPCR had allele frequency at 7.75% and 4.42%, respectively, suggesting they are all positive samples; thus tissue results were false negative. Results described further supported the necessity of liquid biopsy to avoid false negative results from tissue biopsy.
Confirmation of the cut-off value for dPCR as a companion diagnostic tool
The cut-off value was also confirmed through receiver operating characteristic (ROC) curve analysis on the same patient cohort of 1,026 enrolled in this study comparing to ADx-ARMS PCR results. Similar analyses were also performed on 45 sample with paired tissue biopsy, using results from tissue as reference. 0.11% has the highest Youden index for the 1,026 plasma samples, data not shown. Interestingly, higher Youden index were observed when tissue results were used as reference. However, they were below the limit of detection of dPCR, suggesting continuous improvement of assay sensitivity is urgently needed to avoid false negative results. We selected the cut-off value corresponding to the highest Youden Index, which is also at 0.11%.
When the cut off value is set at 0.11%, the sensitivity between the dPCR and ADx-ARMS PCR was 97.53% (95% CI: 93.80–99.32%), the specificity was 89.47% (95% CI: 87.23–91.44%), the PPV was 63.45% (95% CI: 57.14–69.44%), the NPV was 99.49% (95% CI: 98.69–99.86%) and the concordance was 90.74% (95% CI: 88.80–92.44%) which is extremely close to results with cut-off value set at 0.1% as shown in Table 2. Results from 45 paired tissue and plasma samples follow were exactly the same (Table 4). Since cut-off value of 0.1% and 0.11% had very minimal difference in sensitivity and specificity of the assay, we decided to use 0.1% allele frequency as a cut-off value for the assay.
Evaluation of clinical responses of Osimertinib on EGFR p.T790M positive patients tested by digital PCR
In order to further confirm that the additional positive samples determined by dPCR and negative for ADx-ARMS PCR are not false positive samples, the follow-up clinical outcomes were collected, and clinical responses were evaluated according to RECIST 1.1 version. Patients tested positive by either ADx-ARMS PCR and/or dPCR, who received Osimertinib therapy were included in this analysis.
Up to the data cut-off date (Jan. 9th 2021), a total of 75 patients detected as EGFR p.T790M positive received osimertinib treatment, including 74 by dPCR and 1 by ADx-ARMS PCR. Table 5 showed the overall response rate (ORR) and disease control rate (DCR) for dPCR and ADx-ARMS PCR positive patients, respectively. The ORR was 44.59% (33/74, 95% CI: 33.02–56.61%), and the DCR was 90.54% (67/74; 95% CI: 81.48–96.11%) for the dPCR-positive patients. The ORR was 44.90% (22/49; 95% CI: 30.67–59.77%), and the DCR was 91.84% (45/49; 95% CI: 80.40–97.73%) for the ADx-ARMS PCR-positive patients. Notably, for the 26 patients receiving Osimertinib detected as positive by dPCR yet negative by ADx-ARMS PCR, there were still 12 patients experiencing response including 1 with complete response, with the ORR of 46.15% (11/26; 95% CI: 59–66.63%) and DCR of 88.46% (23/26; 95% CI: 69.85–97.55%). These results suggested that dPCR as a companion diagnostic tool is equivalent to ADx-ARMS PCR but more patients would be qualified to use target therapy due to improved test sensitivity.
Table 5
| Best response | dPCR positive | ARMS-PCR positive | dPCR positive/ARMS-PCR negative |
|---|---|---|---|
| CR | 1 | 0 | 1 |
| PR | 32 | 22 | 11 |
| SD | 34 | 23 | 11 |
| PD | 7 | 4 | 3 |
| Total | 74 | 49 | 26 |
| ORR | 44.59% (33.02–56.61%) | 44.90% (30.67–59.77%) | 46.15% (26.59–66.63%) |
| DCR | 90.54% (81.48–96.11%) | 91.84% (80.40–97.73%) | 88.46% (69.85–97.55%) |
EGFR, epidermal growth factor receptor; dPCR, digital polymerase chain reaction; ARMS-PCR, amplification refractory mutation system-polymerase chain reaction; CR, complete response; PR, partial response; SD, stable disease; PD, progressed disease; ORR, overall response rate; DCR, disease control rate.
Discussion
As the presence of EGFR p.T790M is a main cause underlying the development of resistance to 1st and 2nd generation EGFR-TKI treatment, assessment of EGFR p.T790M status plays a decisive role in guiding next-line treatment. Due to tumor heterogeneity, liquid biopsy became extremely important to avoid false negative testing results. There are numerous methods that are available for detecting EGFR p.T790M, including ADx-ARMS PCR, dPCR, NGS, etc. dPCR has the potential to be an ideal biomarker detection method due to its comparatively high sensitivity, fast turnaround time, and easy data interpretation.
The main goal of our study is to evaluate the sensitivity and specificity of the chip-based dPCR assay for clinical use. Because there were no digital PCR assays approved for clinical use on the market that had extensive clinical validation data, a qPCR assay, which was the most similar assay to dPCR, was used. The ADx-ARMS PCR assay was chosen as the comparator assay because it had been extensively validated clinically and had been routinely used in diagnostic labs. For samples that were unconcordant between dPCR and ADx-ARMS PCR, Cobas PCR was used to retest those samples. By testing non-concordant and concordant samples with Cobas PCR, the results showed that it was a better assay than ADx-ARMS PCR but the sensitivity was not as good as dPCR. Using mutant allele frequency determined by dPCR, it also became evident that ADx-ARMS PCR and Cobas PCR had false negative test results due to lack of detection sensitivity at lower allele frequency especially between 0.1% and 1%. This is partly due to the relative quantification of both methods, unlike dPCR which uses absolute quantification, and due to the threshold being set manually for qPCR. For dPCR, positive samples were further confirmed by the mutation status obtained from paired tissue samples, suggesting improved sensitivity did not lead to compromised specificity.
The cut-off value of 0.1% that was used was determined by a previously published study (30). In this study by Zhou et al., they investigated the detection of EGFR p.T790M mutation in NSCLC patients using cell-line samples. One of the criteria for finding a sample mutation positive for dPCR was a mutant allele frequency greater than 0.1%, using the equation cited in the Methods section. This limit of detection was determined using both 20ng and 80ng of input DNA making the criteria applicable for the applications of this paper. To validate the cut-off value, the ROC curve analysis as well as the corresponding Youden Index value was used. While the highest Youden Index value was 0.11%, due to minimal differences between the sensitivity, specificity, PPV, NPV, and concordance between 0.11% and 0.1% cut-off the decided cut-off value for this assay was 0.1% mutant allele frequency.
A previously published study cites that the sensitivity of Cobas PCR is 61% vs. 53.85% from our study when results from paired tissue and plasma samples were compared (31). However, this is not a fair comparison because the input amount of DNA is different between the published study and ours. 50 ng of input DNA Cobas PCR were used in the published study while majority of the samples only had close to 20 ng of cfDNA (32). This means that the limit of detection according to the user manual would be around 23 copies/reaction. Additionally, ADx-ARMS® EGFR mutation detection kit required 10 ng of input DNA per reaction for a limit of detection of 1% mutant allele frequency, or 6–7 copies/reaction (33). Another source did site the LoD for ADx-ARMS PCR ranges from 0.5% to 7.02% depending on the mutation, but which mutation corresponded to what LoD was not described and the input amount of DNA to achieve that LoD was not described as well (34). On the other hand, the dPCR detection kit used 20 ng of input DNA making the limit of detection around 6–7 copies/reaction. Due to the input amount of DNA of the qPCR assays being larger than the dPCR assay, the established limit of detection for the two qPCR assays would not be a fair comparison with dPCR. Additionally, 25 mL of blood was required for the qPCR assays while 10 mL of blood was used in this study. In this study, we used the same amount of input DNA for all assays and found that, although the Cobas PCR assay had better sensitivity than ADx-ARMS PCR, dPCR had the best sensitivity out of all the assays tested. This is most clearly shown when comparing the results of paired tissue and plasma samples. Of the 13 samples determined to be positive by tissue biopsy, dPCR found 7 samples positive while Cobas PCR found 4 samples positive, and ADx-ARMS PCR found 4 samples positive. The samples were confirmed to be true positive because they were compared against tissue biopsy. When the detection of all assays is compared equally, dPCR still has better sensitivity than both assays although the sensitivity was numerically low, at 53.85%.
While dPCR is capable of detecting reproducibly mutant allele frequency of 0.1–1%, it is important for potential users to understand there are upstream sample processing requirements that needs to be strictly followed to maintain the published sensitivity. We believe when a dPCR test is carried out, the following should be taken into consideration: (I) the upstream sample processing should yield enough ctDNA for the test because input amount of ctDNA would directly interfere with the LoD of the assay (30). Our previous study demonstrated that for EGFR p.T790M, 20 ng of ctDNA is required for the LoD to be at 0.1%, which is about 6 genomic copies of the mutant allele. If a lower amount of input DNA is used, the absolute number of copies would further reduce, causing 0.1% LoD is not achievable. (II) The baseline of the assay must be known. dPCR tends to have false positive micro-reactions. We have observed a range of 0−3 false positive wells with 20 ng DNA and 0−4 positive wells with 80 ng DNA. It is important that a positive sample is determined not only by the mutant allele frequency as greater than 0.1% but also by minimal mutant (FAM) positive micro-reactions, to be more than 5. This is effective to avoid false positive test results and will improve the overall accuracy of the assay significantly. (III) Quality control parameters need to be established. For chip based-dPCR, many problems can arise that are beyond the assay itself, which include issues listed above. For instance, a chip can be partially loaded with sample due to problems with the chip loading mechanism. With an improper amount of total wells detected due to sample not reaching a majority of the wells, the number of wells that are positive for the mutation will be skewed, and the results would be deemed unreliable. Therefore, a minimum number of wells detected needs to be established. Another parameter would be to establish a minimum number of reference wells detected as having too few wells would make the number of mutant positive wells unreliable. With the proper precaution in place, the sensitivity and specificity of dPCR can be well maintained to benefit patients better in comparison to assays based on other technologies for companion diagnostics use.
There is another very common method for testing liquid biopsy beyond PCR, which is NGS. NGS has been praised for its ability to broadly detect mutations while PCR can only detect specific mutations (35). This implies that NGS can detect specific mutations as well as new mutations that can arise throughout treatment. Although more mutations detected are of value to drug developer and the research community, even provide additional insights to tumor for research doctors, clinics would still routinely need tests for specific mutations such EGFR mutations as companion diagnostic tools for target therapy. While NGS provided additional mutation information that may not be needed for a simple companion diagnostic test, one drawback with NGS for companion diagnostic is that the LoD is not as great as dPCR (36). A higher LoD results in more false-negatives resulting in patients losing the opportunity for targeted therapy due to false test results. Additionally, NGS does not have the capability of absolute quantification either, making monitoring of changes of mutant allele frequency unreliable (37,38). As demonstrated in the results section, absolute quantification can not only improve the detection sensitivity, but also provide more reliable results for samples with low allele frequencies beyond the detection limit of ARMS-PCR assays, which results in a significant clinical benefit with decreased false positive and false negative rates. With these flaws, as well as the immense cost of NGS due to the complex bioinformatics system and processing speeds needed to make NGS possible, dPCR is more favorable for companion diagnostic use.
Our study has some limitations. First, because the number of paired tissue and plasma samples was limited, it is a great proof of concept to conclude that dPCR is more sensitive than ADx-ARMS PCR without compromising its specificity. More patients need to be enrolled to make a stronger statement statistically. Second, we should identify independently patients who tested positive by dPCR and negative for ADx-ARMS PCR and provide therapy to them. Their response to target therapy should further support the benefit of improved sensitivity from dPCR. Lastly, although dPCR had better sensitivity and overall consistency between plasma and tissue samples test results when compared to ADx-ARMS PCR, it could only detect about 50% of positive samples, suggesting that the sensitivity of liquid biopsy still needs to be improved. This is especially critical for patients with advanced disease and multiple tumor sites, which can make tissue biopsy a challenge or even no longer an option due to inter- and intratumor heterogeneity. In the case of EGFR p.T790M mutation detection, it is mostly acquired resistant mutations; at that stage, most patients would have multiple tumors. Unfortunately, the current state of liquid biopsy does not accommodate all of the needs of these patients.
Conclusions
In conclusion, the newly developed dPCR detection has comparatively high sensitivity without compromising high specificity. dPCR is able to detect lower allele frequencies than ADx-ARMS PCR, and those additional positive results would benefit patients by allowing them to use target therapy to improve OS. Standards and protocols need to be followed vigorously digital PCR to realize its’ improved sensitivity as a liquid biopsy test.
Acknowledgments
Funding: Support for the study was provided by the National key research and development project (2019YFC1315700, to Dr. Jie Wang), the National Natural Sciences Foundation Key Program (81630071, to Dr. Jie Wang); CAMS Innovation Fund for Medical Sciences (CIFMS 2016-I2M-3-008, to Jie Wang; 2017-I2M-1-005, to Dr. Zhijie Wang); Aiyou foundation (KY201701, to Dr. Jie Wang), CAMS Key lab of translational research on lung cancer (2018PT31035, to Dr. Jie Wang), and the National Natural Sciences Foundation (81871889 and 82072586 to Dr. Zhijie Wang) and Beijing Natural Science Foundation (7212084 to Dr. Zhijie Wang); Beijing Hope Run Special Fund of Cancer Foundation of China (LC2020B09 to Dr. Jiachen Xu).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://dx.doi.org/10.21037/tlcr-21-564
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tlcr-21-564
Peer Review File: Available at https://dx.doi.org/10.21037/tlcr-21-564
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tlcr-21-564). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was approved by ethics board of National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College (No. 17-128/1384), ethics board of Shanghai Pulmonary Hospital (No. 108BL, 108BL-1, 108BL-2), ethics board of Affiliated Hospital of Jiangnan University (No. 201 8-002-001), ethics board of Fudan University Shanghai Cancer Center (No. 1804183-1, 1804183-1-1911B), ethics board of Shanghai Chest Hospital (No. LS1821), ethics board of Nanjing Chest Hospital (No. 2018-QL005-01, 2018-QL005-03), ethics board of Peking Union Medical College Hospital (No. HS2018006, KS2018046, KS2019371), ethics board of Tianjin Medical University Cancer Institute & Hospital (No. E219162, E219476), ethics board of First Affiliated Hospital of Soochow University No. 074 (2018), No. 072-2 (2018), ethics board of Tianjin Chest Hospital (No. 2019-001-01, 2019-001-02), ethics board of The Affiliated Cancer Hospital of Nanjing Medical University / Jiangsu Cancer Hospital /Jiangsu Institute of Cancer Research (No. 2019-038, 2019-038-02), ethics board of Zhongshan Hospital [No. 2018-030, 2018-038(2)], ethics board of Peking University Cancer Hospital & Institute (No. 2017TW20-ZY01, 2017TW20-ZY02), ethics board of Tianjin Haihe Hospital [No. 2018HHQX-003, 2018HHQX-003(02)] and ethics board of Tianjin medical university general hospital,/Tianjin Medical University General Hospital (No. IRB2018-101-01, IRB2018-101-03). The general principles of the International Ethical Guidelines for Biomedical Research Involving Human Subjects, the International Conference on Harmonisation guidelines on Good Clinical Practice, and the Declaration of Helsinki (as revised in 2013) were followed. All participants had signed informed consent. The full study protocol was reviewed by all hospital involved in this study and can be accessed via those hospitals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169-81. [Crossref] [PubMed]
- Wang Z, Erb B. Receptors and Cancer. Methods Mol Biol 2017;1652:3-35. [Crossref] [PubMed]
- Yun CH, Boggon TJ, Li Y, et al. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell 2007;11:217-27. [Crossref] [PubMed]
- Wu YL, Zhou C, Hu CP, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. Lancet Oncol 2014;15:213-22. [Crossref] [PubMed]
- Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010;11:121-8. [Crossref] [PubMed]
- Kim ES, Hirsh V, Mok T, et al. Gefitinib versus docetaxel in previously treated non-small-cell lung cancer (INTEREST): a randomised phase III trial. Lancet 2008;372:1809-18. [Crossref] [PubMed]
- Shi Y, Zhang L, Liu X, et al. Icotinib versus gefitinib in previously treated advanced non-small-cell lung cancer (ICOGEN): a randomised, double-blind phase 3 non-inferiority trial. Lancet Oncol 2013;14:953-61. [Crossref] [PubMed]
- Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013;31:3327-34. [Crossref] [PubMed]
- Lin Y, Wang X, Jin H. EGFR-TKI resistance in NSCLC patients: mechanisms and strategies. Am J Cancer Res 2014;4:411-35. [PubMed]
- Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A 2008;105:2070-5. [Crossref] [PubMed]
- Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res 2013;19:2240-7. [Crossref] [PubMed]
- Sharma SV, Lee DY, Li B, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010;141:69-80. [Crossref] [PubMed]
- Ohashi K, Maruvka YE, Michor F, et al. Epidermal growth factor receptor tyrosine kinase inhibitor-resistant disease. J Clin Oncol 2013;31:1070-80. [Crossref] [PubMed]
- Cross DA, Ashton SE, Ghiorghiu S, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov 2014;4:1046-61. [Crossref] [PubMed]
- Greig SL. Osimertinib: First Global Approval. Drugs 2016;76:263-73. [Crossref] [PubMed]
- Ramalingam SS, Vansteenkiste J, Planchard D, et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N Engl J Med 2020;382:41-50. [Crossref] [PubMed]
- Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689-99. [Crossref] [PubMed]
- Goss G, Tsai CM, Shepherd FA, et al. Osimertinib for pretreated EGFR Thr790Met-positive advanced non-small-cell lung cancer (AURA2): a multicentre, open-label, single-arm, phase 2 study. Lancet Oncol 2016;17:1643-52. [Crossref] [PubMed]
- Mok TS, Wu Y-L, Ahn M-J, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Wang Z, Cheng Y, An T, et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): a phase 2, single-arm, multicentre clinical trial. Lancet Respir Med 2018;6:681-90. [Crossref] [PubMed]
- Duan J, Xu J, Wang Z, et al. Refined Stratification Based on Baseline Concomitant Mutations and Longitudinal Circulating Tumor DNA Monitoring in Advanced EGFR-Mutant Lung Adenocarcinoma Under Gefitinib Treatment. J Thorac Oncol 2020;15:1857-70. [Crossref] [PubMed]
- Xu T, Kang X, You X, et al. Cross-Platform Comparison of Four Leading Technologies for Detecting EGFR Mutations in Circulating Tumor DNA from Non-Small Cell Lung Carcinoma Patient Plasma. Theranostics 2017;7:1437-46. [Crossref] [PubMed]
- Whale AS, Huggett JF, Cowen S, et al. Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acids Res 2012;40:e82. [Crossref] [PubMed]
- Nixon G, Garson JA, Grant P, et al. Comparative study of sensitivity, linearity, and resistance to inhibition of digital and nondigital polymerase chain reaction and loop mediated isothermal amplification assays for quantification of human cytomegalovirus. Anal Chem 2014;86:4387-94. [Crossref] [PubMed]
- Zonta E, Garlan F, Pécuchet N, et al. Multiplex Detection of Rare Mutations by Picoliter Droplet Based Digital PCR: Sensitivity and Specificity Considerations. PLoS One 2016;11:e0159094. [Crossref] [PubMed]
- Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015;90:509-15. [Crossref] [PubMed]
- Feng Q, Gai F, Sang Y, et al. A comparison of QuantStudio™ 3D Digital PCR and ARMS-PCR for measuring plasma EGFR T790M mutations of NSCLC patients. Cancer Manag Res 2018;10:115-21. [Crossref] [PubMed]
- Zhou RF CY, Li ZL, et al. A digital PCR assay development to detect EGFR T790M mutation in NSCLC patients. Frontiers in Laboratory Medicine 2018;2:89-96. [Crossref]
- Jenkins S, Yang JC, Ramalingam SS, et al. Plasma ctDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:1061-70. [Crossref] [PubMed]
- Roche Diagnostics. cobas® EGFR Mutation Test v2: For in vitro diagnostic use. 2016;v2:60.
- Amoy Diagnostics. Human EGFR gene mutation detection kit (fluorescent PCR method) manual. 2015;P3.3:2.
- Walsh K, Wallace WA. Molecular pathology in lung cancer: a guide to the techniques used in clinical practice. Histopathology 2014;65:731-41. [Crossref] [PubMed]
- Bos MK, Nasserinejad K, Jansen MPHM, et al. Comparison of variant allele frequency and number of mutant molecules as units of measurement for circulating tumor DNA. Mol Oncol 2021;15:57-66. [Crossref] [PubMed]
- Bartels S, Persing S, Hasemeier B, et al. Molecular Analysis of Circulating Cell-Free DNA from Lung Cancer Patients in Routine Laboratory Practice: A Cross-Platform Comparison of Three Different Molecular Methods for Mutation Detection. J Mol Diagn 2017;19:722-32. [Crossref] [PubMed]
- Forghieri F, Comoli P, Marasca R, et al. Minimal/Measurable Residual Disease Monitoring in NPM1-Mutated Acute Myeloid Leukemia: A Clinical Viewpoint and Perspectives. Int J Mol Sci 2018;19:3492. [Crossref] [PubMed]
- Takamatsu H, Wee RK, Zaimoku Y, et al. A comparison of minimal residual disease detection in autografts among ASO-qPCR, droplet digital PCR, and next-generation sequencing in patients with multiple myeloma who underwent autologous stem cell transplantation. Br J Haematol 2018;183:664-8. [Crossref] [PubMed]


