
A multiethnic bidirectional Mendelian randomization study negates causal effects of C-reactive protein concentrations on lung cancer
Introduction
Cancer is a leading global public health issue and the second major cause of death in the United States. Particularly, lung cancer (LC) is currently the second most common cancer and the major cause of cancer-related death globally (1). The American Cancer Society estimates that 235,760 new LC cases and 131,880 LC deaths are projected to occur in the United States in 2021 (2). Thus, early recognition of potentially modifiable risk factors contributes to better prevention of LC.
Growing studies have demonstrated that chronic inflammation, especially within the respiratory system, might be a risk factor for LC (3-6). Several inflammatory conditions, such as chronic pulmonary infection (7) and chronic obstructive pulmonary disease (8), were correlated with an increased risk of LC. It has been proposed that LC could be induced by pulmonary inflammation through several pathways, such as the increase in angiogenesis during the repair of the damaged tissue and the generation of reactive oxygen (9). Simultaneously, circulating C-reactive protein (CRP) was considered as a systemic marker of chronic inflammation (10), and elevated CRP blood concentrations have been widely evaluated for their association with LC (3,11-20). Up till now, two assumptions have been presented to explain the mechanisms between elevated CRP concentrations and the risk of LC (10). On the one hand, elevated CRP concentrations and chronic inflammation are causally related to the carcinogenesis of the lung indeed. On the other hand, CRP concentrations may also indicate a systemic response to ongoing disease or comorbidity, such as an occult cancer or a premalignant state. The majority of previous cohort studies and a meta-analysis (21) denoted that elevated CRP concentrations were correlated with LC occurrence. However, given that cigarette smoking is by far the leading risk factor for LC, CRP is widely seen as a predictor of LC in smokers and former smokers (22), while cigarette smoking itself has been reported to increase circulating levels of CRP directly (23). Hence, conventional observational studies are prone to bias by potential confounding factors or reverse causation, and the causality was underpowered for definitive conclusions from previous studies.
Using genetic variations as instrumental variables (IVs), Mendelian randomization (MR) analysis is a novel epidemiology method to estimate the causation between an exposure and an outcome, with less impressionability to reverse causation and unmeasured confounders. In settings for which the IV assumptions are well justified, the findings could help optimize drug development or clinical trials and inform clinical or public health decision-making (24). Single nucleotide polymorphism (SNP) is a single base-pair difference in the DNA sequence of individuals within a species, which is the most common type of genetic variation in humans (25). Given that genetic variants are allocated at conception randomly, they are generally independent of environmental risk factors and precede the risk factors and the diseases’ onset (26). As for CRP, it has been estimated that the heritability of CRP blood concentrations was estimated from 25% to 40% (27), suggesting that genetic architecture might contribute to modulating CRP concentrations. Meanwhile, based on the summary data from genome-wide association studies (GWAS), our two-sample MR analysis could assess their causal relationships more comprehensively with robust statistical power (28,29). Previous MR studies (5,30-32) have illustrated that no causation was detected between genetically regulated elevated CRP concentrations and risk of LC. However, the sample size of LC patients in previous MR studies was relatively limited, insufficient to provide adequate statistical power to evaluate their causal nexus. Then, due to the limitation of the GWASs at the time, causal inferences provided by previous studies solely applied to populations of European ancestry; therefore, the findings were lack of generalizable value. Moreover, since genetically higher LC risk can influence CRP concentrations, no bidirectional MR study has been conducted, and therefore, previous findings were likely to be biased by reverse causality.
Generally, using the recent largescale meta-analysis of the GWASs updating CRP-specific SNPs among diverse populations, the present study could offer the latest comprehensive evidence for assessing the bidirectional causal effect between genetically regulated CRP concentrations and risk of LC with the two-sample MR method.
Methods
Study participants of lung cancer
In general, 11,348 LC cases and 15,861 controls from the International Lung Cancer Consortium (ILCCO) (33), an international research group that established and implemented LC research from different geographical areas and ethnicities, were used as epidemiological summary-level data. Subgroup analyses were stratified according to the pathology classification of non-small cell lung cancer (NSCLC). Specifically, we divided types of cancer into lung adenocarcinoma (LUAD) (3442 cases, 14,894 controls) as well as squamous cell lung cancer (LUSC) (3275 cases, 15,038 controls). The study was conducted following the Declaration of Helsinki (as revised in 2013) (34).
Genetic variants associated with elevated CRP concentrations
The latest large-scale GWAS datasets were retrieved from MR-Base, an open GWAS project developed at the MRC Integrative Epidemiology Unit (IEU) at the University of Bristol (35). Through Bonferroni correction to control the family-wise error rate (FWER), the P=5×10−8 threshold was widely accepted for association identification between a common genetic variant and a trait of interest, given the linkage disequilibrium (LD) structure of the genome (36). Using the MR-Base platform, SNPs associated with elevated CRP concentrations were initially selected from Neale Lab, Pan-UK Biobank (UKB) team, RIKEN Center for Integrative Medical Sciences, and the European Bioinformatics Institute (EBI) database of complete GWAS summary data at the genome-wide significance threshold (P<5×10-8). Utilizing LD analysis, we attempted to exclude SNPs once mutual LD surpassed the limited value (R2<0.001) with a larger P value conjugately. Eventually, the final IV set was established, including 204 SNPs (available online: https://cdn.amegroups.cn/static/public/tlcr-21-750-01.xlsx). Among them, 174 SNPs were of European ancestry, 13 SNPs were of Hispanic or Latin American origin, 7 SNPs were of South Asian ancestry, 6 SNPs were of East Asian ancestry, and 4 SNPs were of African American or Afro-Caribbean ancestry. Risk alleles and baseline alleles were encoded separately in accordance with the association with the rate of CRP concentrations. As the sample size of 204 instruments and 310,305 individual samples in our study, the F-statistic was 3,135.39 as estimated (37) given the type I error rate =0.05, which suggested noticeable correlativity for the MR analysis.
Statistical analysis
MR method was implied as the statistical analysis tool, which possesses three suppositions as foundations (38): (I) the IVs are robustly correlating with elevated CRP concentrations; (II) the IVs affect LC merely via their effect on elevated CRP concentrations (i.e., the IVs are independent of the outcome given the exposure), and (III) the IVs are independent of any confounders. As demonstrated by the previous studies (35) and the screening procedure of SNPs described in the previous paragraph, the first assumption was met.
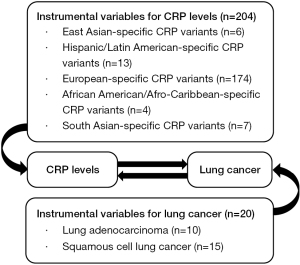
Two-sample MR analyses were conducted to evaluate the potential bidirectional associations between elevated CRP concentrations and LC risk (Figure 1). In addition to the aggregate effect, causal inferences were obtained from five independent ancestries for specific evaluation among different ethnic groups. Five well-established MR analytical methods were applied, including inverse-variance weighted (IVW), weighted median, MR-Egger, weighted mode, and simple mode. IVW was applied to the combination of multiple IVs as sole estimation of genetic variants by weighted score, which has the most statistical power among the five methods (39). The rest of the methods were performed for sensitivity analysis and to indirectly test the second assumption. Global pleiotropic effects were obtained from the MR-Egger analyses based on the intercept. Cochran's Q statistic and I2 were inspected to estimate heterogeneity. Furthermore, we conducted leave-one-out analyses to investigate whether the estimation of MR was determined or biased by an individual SNP by omitting a single SNP successively. MR analyses were performed in R (version 3.6.2) using the package TwoSampleMR (version 0.5.0) (35).
Results
The basic characteristics and their F statistics for selected summary level GWASs applied in our MR study are given in Table 1, and the detailed information of each SNP used in our study is listed in table available online: https://cdn.amegroups.cn/static/public/tlcr-21-750-01.xlsx.
Table 1
| Trait | Sample size | Sex | First author/consortium | F statistics |
|---|---|---|---|---|
| Overall C-reactive protein (CRP) | 310,305 | Males and females | – | 3,135.39 |
| East Asian-specific CRP variants | 75,391 | Males and females | Ishigaki K | 762.53 |
| Hispanic/Latin American-specific CRP variants | 15,912 | Males and females | Wojcik GL | 161.73 |
| European-specific CRP variants | 204,402 | Males and females | Neale lab | 2,065.67 |
| African American/Afro-Caribbean-specific CRP variants | 6,203 | Males and females | Pan-UKB team | 63.66 |
| South Asian-specific CRP variants | 8,397 | Males and females | Pan-UKB team | 85.82 |
| Lung cancer | 27,209 | Males and females | ILCCO | 275.84 |
| Lung adenocarcinoma | 18,336 | Males and females | ILCCO | 186.21 |
| Squamous cell lung cancer | 18,313 | Males and females | ILCCO | 185.98 |
ILCCO, the International Lung Cancer Consortium.
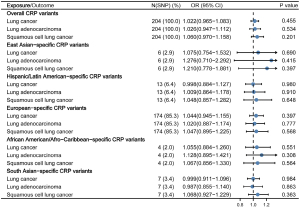
In the MR analysis of mixed ancestries using 204 SNPs, the outcome of IVW method demonstrated that although genetically regulated one-unit increased in the log-transformed CRP concentrations were associated with a relatively 2.2% increased risk of LC, no statistically meaningful conclusions could be drawn (OR =1.022, 95% CI: 0.965–1.083, P=0.455) (Figure 2), which was also consistent regarding pathological subtypes including LUAD (OR =1.026, 95% CI: 0.947–1.112, P=0.534) and LUSC (OR =1.060, 95% CI: 0.970–1.158, P=0.201). Findings were consistent among sensitivity analyses, demonstrating null causal relationship between both phenotypes (available online: https://cdn.amegroups.cn/static/public/tlcr-21-750-02.xlsx). Meanwhile, as for the causal effect among different ethnic populations, no association reached statistical significance among East Asian (OR =1.075, 95% CI: 0.754–1.532, P=0.690), Hispanic/Latin American (OR =0.998, 95% CI: 0.884–1.127, P=0.980), European (OR =1.044, 95% CI: 0.945–1.155, P=0.397), African American/Afro-Caribbean (OR =1.055, 95% CI: 0.884–1.260, P=0.551), and South Asian population (OR =0.999, 95% CI: 0.911–1.096, P=0.984). Detailed results on LC subtypes of ethnic-specific analyses are presented in Figure 3.
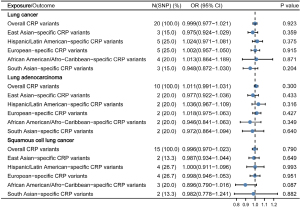
Regarding the impact of LC upon CRP concentrations, similar modest and nonsignificant estimates of causal effect were observed (OR =0.999, 95% CI: 0.977–1.021, P=0.923), conforming the absence of bidirectional effects. Subtypes including LUSC (OR =0.996, 95% CI: 0.970–1.023, P=0.790) and LUAD (OR =1.011, 95% CI: 0.991–1.031, P=0.300) supported the same conclusion. Lack of causal association also predominated among population subgroups (Figure 3).
The results of sensitivity analyses among all study populations are shown in table available online: https://cdn.amegroups.cn/static/public/tlcr-21-750-02.xlsx. Using MR-Egger regression, our findings did not support the existence of global pleiotropic assumptions in most of the study outcomes (Table S1), while evidence of heterogeneity was found in LC effect on European-specific CRP variants (Table S2). Leave-one-out studies of the LC overall and subgroup analyses showed no evidence that a single SNP had an impact upon the overall effect of CRP variants on LC risks (available online: https://cdn.amegroups.cn/static/public/tlcr-21-750-03.xlsx).
Discussion
This is the first study to conduct bidirectional MR analyses between CRP concentrations and LC among different ethnic backgrounds. No bidirectional causation between genetically regulated elevated concentrations of CRP and LC risk was observed in our study among East Asian (P=0.690, nSNP =6, n=75,391), Hispanic/Latin American (P=0.980, nSNP =13, n=15,912), European (P=0.397, nSNP =174, n=204,402), African American/Afro-Caribbean (P=0.551, nSNP =4, n=6,203), and South Asian populations (P=0.984, nSNP =7, n=8,397). Additionally, no causality was found in subgroup analyses concerning pathologic types as well.
The results are inconsistent with most previous prospective cohort studies (3,11,12,14,16-19) using measured plasma concentrations of circulating CRP to explore the risk of LC. Shiels et al. (16) demonstrated that elevated measured CRP concentrations were related to the risk of LC in two independent studies from the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. Allin et al. (11) also illustrated that elevated plasma concentrations of CRP in cancer-free individuals were relevant to an increased risk of LC. Besides, the latest meta-analysis (21) denoted that elevated plasma concentrations of CRP were also related to an increased risk of LC. Interestingly, after a stratified analysis (15) based on smoking status, the positive association between circulating high sensitivity CRP concentrations and the risk of LC disappeared when considering non-smokers. Thus, rather than a causal factor, the authors (15) concluded that circulating high sensitivity CRP concentration could be a pre-diagnostic marker of LC. Since traditional observational studies are vulnerable to potential confounding factors or reverse causation, we supposed that previous results might be influenced by underlying confounders (e.g., cigarette smoking status and older age stratifications).
Our study showed similar results in effect directions compared with previous MR studies (5,30-32), whereas these studies came to the conclusions from a limited LC sample size. Among them, the largest sample size of LC patients was only 416 (30). Therefore, they might not be precise enough to evaluate causation between genetically elevated CRP concentrations and LC risk due to the deficiency of statistical power. In comparison, we specifically selected a total of 27,209 participants from ILCCO with 11,348 LC cases and 15,861 controls as our study participants, which were much more extensive than previous ones and offered adequate statistical power. Meanwhile, two (5,30) of them didn’t identify SNPs through GWAS, and the majority of chosen SNPs haven’t been confirmed by subsequent GWAS (27,40,41), indicating those SNPs might not be authentically associated with elevated CRP concentrations. Even for the other two studies (31,32) with GWAS-identified SNPs, some of their chosen SNPs were also proposed to have nothing to do with elevated CRP concentrations subsequently (27,40,41), such as rs4903031, rs6901250, and rs4705952. Additionally, genetic pleiotropy could occur without sensitivity analyses, indicating previously chosen SNPs might relate to other inflammatory processes and result in a spurious association. Consequently, biased estimation for the causation could happen due to those irrelevant SNPs and potential global pleiotropic effects in previous MR studies. Furthermore, more relevant SNPs have been updated in recent years. Our study included 204 SNPs from the latest GWAS as the IV set, which would explain the variance of elevated CRP concentrations more comprehensively and precisely. Generally, our updated MR study provided the latest and much more substantial evidence for evaluating the causality between genetically regulated elevated CRP concentrations and LC risk.
Regarding the etiology of LC, increasing evidence have illustrated that chronic inflammation may play a significant role amid the carcinogenesis process (4-7,16,42,43). Inflammatory conditions, such as chronic obstructive pulmonary disease (3,44-47), chlamydia pneumonia (4,48), tuberculosis (6), and human immunodeficiency virus (HIV) (49) seem to correlate with an increased LC risk. Through the nuclear factor kappa B (NF-κB) pathway, chronic inflammation may actively promote the malignant process, the increase in angiogenesis and proliferation of cells during tissue repair, and the up-regulation of antiapoptotic genes through reactive oxygen and nitrogen species production (9). Considered a systemic marker of chronic inflammation (5), elevated plasma CRP concentrations were associated with an increased LC risk in many conventional observational studies. However, our MR analysis denoted that elevated CRP concentrations were not causally associated with LC risk, which could be explained from two aspects. For one thing, chronic inflammation promotes the carcinogenesis of the lung, possibly by means of other inflammatory mediators except for CRP. It has been reported that soluble tumor necrosis factor receptor-2 (sTNFRII), serum amyloid A (SAA), and monokine induced by gamma interferon (CXCL9/MIG) were also related to LC risk (16). For another, due to the natural defects of observational studies, their results could be influenced by unmeasured confounders, such as smoking status and elder age. In addition, inflammation and elevated CRP concentrations could be induced by tumor growth, and CRP could also be an indicator of an immune response to tumor antigens (10). Consequently, elevated CRP concentrations might also respond to occult or very early-stage cancer (16). However, CRP may not always act as a cancer promoter but sometimes play a role in treating cancer. Sasaki et al. observed that CRP can inhibit lymph node metastasis and lymphatic angiogenesis of squamous cell carcinoma through injecting CRP into mice model (50). As for the prognostic effects of CRP, Shinohara and colleagues found high serum circulating CRP levels on postoperative days were associated with enhanced 5-year overall survival (OS) as well as recurrence-free survival (RFS) in NSCLC patients (51). Instead, Okada et al. reported the high serum CRP levels during perioperative period were a poor prognostic factor for OS and RFS in NSCLC patients (52). Consequently, the impact of CRP levels on the incidence and prognosis of LC was nuanced and complex, and further studies with larger sample sizes and better designs are in an urgent need.
Several merits deserve explicit mention. First, this is the first study to assess the bidirectional causation between elevated CRP concentrations and LC risk to date. Second, we used the most comprehensive GWAS-identified SNPs as an IV set. Compared with the previous studies, our study with 204 SNPs explained more variance of elevated CRP concentrations. Thanks to the robustly associated IVs (F-statistics =3,135.39) and the great sample size (n=310,305), our study was capable of providing a relatively accurate evaluation of a causal inference. Third, to the best of our knowledge, this is the first MR study that conducted stratification analyses according to different ethnic populations. Thus, we were able to investigate whether their association could vary under different ancestry backgrounds.
Notwithstanding the advantages, several limitations in our study cannot be neglected. First, even though the most comprehensive set of multiethnic genetic variants were applied for our research so far, only a part of the variance of elevated CRP concentrations could be explained in the population. Some unknown CRP-related SNPs, which deserve further investigation in future GWASs, could play an essential role in LC development. Besides, due to the methodology limitations currently, MR assumptions could not be thoroughly tested, and therefore potential violations against the assumptions may occur. To overcome this difficulty, instead of directivity evaluated in the second assumption, we implemented additional sensitivity analyses, which showed the absence of pleiotropic effects in our study, suggesting no violation of the second MR assumption. Moreover, depending on GWAS summary statistics, the two-sample MR methods assume a linear relationship between the exposure and the outcome. Hence, it is possible that a few genetic variants were also related to confounding factors of CRP and LC.
Overall, our study negates bidirectional causal effects of CRP concentrations on LC among East Asian, Hispanic/Latin American, European, African American/Afro-Caribbean, and South Asian populations. Still, it opens a new concept for the current research orientation. Further investigation of the profound relationship between both phenotypes is required to be unveiled in pathologic and biochemistry aspects.
Acknowledgments
Funding: This work was supported by the National Key R&D Program of China (2016YFC0905400), China National Science Foundation (81871893, 81501996), Cultivation of Guangdong College Students’ Scientific and Technological Innovation (“Climbing Program” Special Funds) (Grant number pdjh2020a0480 and pdjh2021a0407), Key Project of Guangzhou Scientific Research Project (201804020030), High-level university construction project of Guangzhou Medical University (20182737, 201721007, 201715907, 2017160107), IVATS National key R&D Program (2017YFC0907903, 2017YFC0112704) and Application, Industrialization and Generalization of Surgical Incision Protector (2011B090400589).
Footnote
Peer Review File: Available at https://dx.doi.org/10.21037/tlcr-21-750
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tlcr-21-750). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted following the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. Erratum in: CA Cancer J Clin 2021;71:359. [Crossref] [PubMed]
- Allin KH, Bojesen SE, Johansen JS, et al. Cancer risk by combined levels of YKL-40 and C-reactive protein in the general population. Br J Cancer 2012;106:199-205. [Crossref] [PubMed]
- Chaturvedi AK, Gaydos CA, Agreda P, et al. Chlamydia pneumoniae infection and risk for lung cancer. Cancer Epidemiol Biomarkers Prev 2010;19:1498-505. [Crossref] [PubMed]
- Chaturvedi AK, Caporaso NE, Katki HA, et al. C-reactive protein and risk of lung cancer. J Clin Oncol 2010;28:2719-26. [Crossref] [PubMed]
- Engels EA, Wu X, Gu J, et al. Systematic evaluation of genetic variants in the inflammation pathway and risk of lung cancer. Cancer Res 2007;67:6520-7. [Crossref] [PubMed]
- Shiels MS, Albanes D, Virtamo J, et al. Increased risk of lung cancer in men with tuberculosis in the alpha-tocopherol, beta-carotene cancer prevention study. Cancer Epidemiol Biomarkers Prev 2011;20:672-8. [Crossref] [PubMed]
- Koshiol J, Rotunno M, Consonni D, et al. Chronic obstructive pulmonary disease and altered risk of lung cancer in a population-based case-control study. PLoS One 2009;4:e7380. [Crossref] [PubMed]
- Ballaz S, Mulshine JL. The potential contributions of chronic inflammation to lung carcinogenesis. Clin Lung Cancer 2003;5:46-62. [Crossref] [PubMed]
- Heikkilä K, Ebrahim S, Lawlor DA. A systematic review of the association between circulating concentrations of C reactive protein and cancer. J Epidemiol Community Health 2007;61:824-33. [Crossref] [PubMed]
- Allin KH, Bojesen SE, Nordestgaard BG. Baseline C-reactive protein is associated with incident cancer and survival in patients with cancer. J Clin Oncol 2009;27:2217-24. [Crossref] [PubMed]
- Allin KH, Nordestgaard BG. Elevated C-reactive protein in the diagnosis, prognosis, and cause of cancer. Crit Rev Clin Lab Sci 2011;48:155-70. [Crossref] [PubMed]
- dos Santos Silva I, De Stavola BL, Pizzi C, et al. Circulating levels of coagulation and inflammation markers and cancer risks: individual participant analysis of data from three long-term cohorts. Int J Epidemiol 2010;39:699-709. [Crossref] [PubMed]
- Il'yasova D, Colbert LH, Harris TB, et al. Circulating levels of inflammatory markers and cancer risk in the health aging and body composition cohort. Cancer Epidemiol Biomarkers Prev 2005;14:2413-8. [Crossref] [PubMed]
- Muller DC, Larose TL, Hodge A, et al. Circulating high sensitivity C reactive protein concentrations and risk of lung cancer: nested case-control study within Lung Cancer Cohort Consortium. BMJ 2019;364:k4981. [Crossref] [PubMed]
- Shiels MS, Katki HA, Hildesheim A, et al. Circulating Inflammation Markers, Risk of Lung Cancer, and Utility for Risk Stratification. J Natl Cancer Inst 2015;107:djv199. [Crossref] [PubMed]
- Shiels MS, Pfeiffer RM, Hildesheim A, et al. Circulating inflammation markers and prospective risk for lung cancer. J Natl Cancer Inst 2013;105:1871-80. [Crossref] [PubMed]
- Siemes C, Visser LE, Coebergh JW, et al. C-reactive protein levels, variation in the C-reactive protein gene, and cancer risk: the Rotterdam Study. J Clin Oncol 2006;24:5216-22. [Crossref] [PubMed]
- Van Hemelrijck M, Holmberg L, Garmo H, et al. Association between levels of C-reactive protein and leukocytes and cancer: three repeated measurements in the Swedish AMORIS study. Cancer Epidemiol Biomarkers Prev 2011;20:428-37. [Crossref] [PubMed]
- Heikkilä K, Harris R, Lowe G, et al. Associations of circulating C-reactive protein and interleukin-6 with cancer risk: findings from two prospective cohorts and a meta-analysis. Cancer Causes Control 2009;20:15-26. [Crossref] [PubMed]
- Guo YZ, Pan L, Du CJ, et al. Association between C-reactive protein and risk of cancer: a meta-analysis of prospective cohort studies. Asian Pac J Cancer Prev 2013;14:243-8. [Crossref] [PubMed]
- Tonstad S, Cowan JL. C-reactive protein as a predictor of disease in smokers and former smokers: a review. Int J Clin Pract 2009;63:1634-41. [Crossref] [PubMed]
- Gallus S, Lugo A, Suatoni P, et al. Effect of Tobacco Smoking Cessation on C-Reactive Protein Levels in A Cohort of Low-Dose Computed Tomography Screening Participants. Sci Rep 2018;8:12908. [Crossref] [PubMed]
- Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 2018;362:k601. [Crossref] [PubMed]
- Schork NJ, Fallin D, Lanchbury JS. Single nucleotide polymorphisms and the future of genetic epidemiology. Clin Genet 2000;58:250-64. [Crossref] [PubMed]
- Lawlor DA, Harbord RM, Sterne JA, et al. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 2008;27:1133-63. [Crossref] [PubMed]
- Dehghan A, Dupuis J, Barbalic M, et al. Meta-analysis of genome-wide association studies in >80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation 2011;123:731-8. [Crossref] [PubMed]
- Burgess S, Scott RA, Timpson NJ, et al. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol 2015;30:543-52. [Crossref] [PubMed]
- Pierce BL, Burgess S. Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am J Epidemiol 2013;178:1177-84. [Crossref] [PubMed]
- Allin KH, Nordestgaard BG, Zacho J, et al. C-reactive protein and the risk of cancer: a mendelian randomization study. J Natl Cancer Inst 2010;102:202-6. [Crossref] [PubMed]
- Heikkilä K, Silander K, Salomaa V, et al. C-reactive protein-associated genetic variants and cancer risk: findings from FINRISK 1992, FINRISK 1997 and Health 2000 studies. Eur J Cancer 2011;47:404-12. [Crossref] [PubMed]
- Prizment AE, Folsom AR, Dreyfus J, et al. Plasma C-reactive protein, genetic risk score, and risk of common cancers in the Atherosclerosis Risk in Communities study. Cancer Causes Control 2013;24:2077-87. [Crossref] [PubMed]
- Wang Y, McKay JD, Rafnar T, et al. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat Genet 2014;46:736-41. [Crossref] [PubMed]
- World Medical Association. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA 2013;310:2191-4. [Crossref] [PubMed]
- Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7:34408. [Crossref] [PubMed]
- Chen Z, Boehnke M, Wen X, et al. Revisiting the genome-wide significance threshold for common variant GWAS. G3 (Bethesda) 2021;11:jkaa056.
- Burgess S, Thompson SGCRP CHD Genetics Collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol 2011;40:755-64. [Crossref] [PubMed]
- VanderWeele TJ, Tchetgen Tchetgen EJ, Cornelis M, et al. Methodological challenges in mendelian randomization. Epidemiology 2014;25:427-35. [Crossref] [PubMed]
- Noyce AJ, Kia DA, Hemani G, et al. Estimating the causal influence of body mass index on risk of Parkinson disease: A Mendelian randomisation study. PLoS Med 2017;14:e1002314. [Crossref] [PubMed]
- Ligthart S, Vaez A, Võsa U, et al. Genome Analyses of >200,000 Individuals Identify 58 Loci for Chronic Inflammation and Highlight Pathways that Link Inflammation and Complex Disorders. Am J Hum Genet 2018;103:691-706. [Crossref] [PubMed]
- Kocarnik JM, Pendergrass SA, Carty CL, et al. Multiancestral analysis of inflammation-related genetic variants and C-reactive protein in the population architecture using genomics and epidemiology study. Circ Cardiovasc Genet 2014;7:178-88. [Crossref] [PubMed]
- Pine SR, Mechanic LE, Enewold L, et al. Increased levels of circulating interleukin 6, interleukin 8, C-reactive protein, and risk of lung cancer. J Natl Cancer Inst 2011;103:1112-22. [Crossref] [PubMed]
- Shiels MS, Engels EA, Shi J, et al. Genetic variation in innate immunity and inflammation pathways associated with lung cancer risk. Cancer 2012;118:5630-6. [Crossref] [PubMed]
- Santillan AA, Camargo CA Jr, Colditz GA. A meta-analysis of asthma and risk of lung cancer (United States). Cancer Causes Control 2003;14:327-34. [Crossref] [PubMed]
- Sin DD, Man SF. Impact of cancers and cardiovascular diseases in chronic obstructive pulmonary disease. Curr Opin Pulm Med 2008;14:115-21. [Crossref] [PubMed]
- Purdue MP, Gold L, Järvholm B, et al. Impaired lung function and lung cancer incidence in a cohort of Swedish construction workers. Thorax 2007;62:51-6. [Crossref] [PubMed]
- Brody JS, Spira A. State of the art. Chronic obstructive pulmonary disease, inflammation, and lung cancer. Proc Am Thorac Soc 2006;3:535-7. [Crossref] [PubMed]
- Littman AJ, Jackson LA, Vaughan TL. Chlamydia pneumoniae and lung cancer: epidemiologic evidence. Cancer Epidemiol Biomarkers Prev 2005;14:773-8. [Crossref] [PubMed]
- Engels EA, Brock MV, Chen J, et al. Elevated incidence of lung cancer among HIV-infected individuals. J Clin Oncol 2006;24:1383-8. [Crossref] [PubMed]
- Sasaki T, Motoyama S, Sato Y, et al. C-reactive protein inhibits lymphangiogenesis and resultant lymph node metastasis of squamous cell carcinoma in mice. Surgery 2013;154:1087-92. [Crossref] [PubMed]
- Shinohara S, Sugaya M, Onitsuka T, et al. Prognostic Impact of Postoperative C-reactive Protein for Non-small Cell Lung Cancer Following Lobectomy. Anticancer Res 2018;38:3193-8. [PubMed]
- Okada S, Shimomura M, Tsunezuka H, et al. Prognostic Significance of Perioperative C-Reactive Protein in Resected Non-Small Cell Lung Cancer. Semin Thorac Cardiovasc Surg 2020;32:1046-55. [Crossref] [PubMed]


