
Malignant thoracic lymph node classification with deep convolutional neural networks on real-time endobronchial ultrasound (EBUS) images
Introduction
Thoracic lymph node (LN) evaluation is essential for the accurate diagnosis of lung cancer and deciding the appropriate course of treatment. Endobronchial ultrasound-guided transbronchial needle aspiration (EBUS-TBNA) is considered a standard method for confirming the presence of LN metastases due to its minimal invasiveness and high accuracy. Endobronchial ultrasound (EBUS) visualization should be performed systematically from nodal stations N3 to N1 (1), and a biopsy should be performed for any LN with a short axis greater than 5 mm. EBUS image assessment not only helps in selecting which LNs to perform a biopsy on, but also aids clinical judgment when the biopsy result is inconclusive (2). Generally, ultrasounds are the most widely used imaging modality among screening and diagnostic tools, owing to their safety, non-invasiveness, cost-effectiveness, and real-time display. However, compared to other imaging modalities, ultrasounds have a relatively low imaging quality due to noise, artifacts and high inter-and intra-observer variability. As for the EBUS, bronchoscopists utilize ultrasonographic features to identify malignant LNs. However, its predictive power is limited by the experience and subjective interpretation of the bronchoscopist.
Recently, the method of using ultrasound imaging analysis with deep learning algorithms has been developed to perform more objective and comprehensive assessments (3). Only a few studies have focused on EBUS prediction models using convolutional neural networks (CNNs) (4). EBUS images should be interpreted in real-time during the EBUS-TBNA procedure to determine whether to perform a biopsy. However, to our knowledge, no previous study has proposed a network that processes dozens of images per second on a single mainstream graphics processing unit (GPU) device to achieve real-time EBUS image analysis.
The aim of this study is to build effective deep CNNs for the automatic classification of malignancy in thoracic LNs using real-time EBUS images. Selecting LNs that require biopsy through real-time EBUS image analysis in conjunction with deep learning is expected to shorten the EBUS-TBNA procedure time, increase the accuracy of nodal staging, and ultimately improve patient safety.
We present the following article in accordance with the STARD reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-21-870/rc).
Methods
Data collection and preparation
Patients with suspected lung cancer, or other malignancies with thoracic LN enlargement, who underwent an EBUS-TBNA procedure to evaluate their thoracic LNs were enrolled from Oct 2019 to Jul 2020, at the Severance Hospital in Seoul, Republic of Korea. The EBUS-TBNA procedure was conducted by a bronchoscopist with a convex probe EBUS (BF-UC260FW; Olympus Co., Tokyo, Japan), and the EBUS images were generated using an ultrasound processor (EU-ME2; Olympus Co.; Figure 1A). All procedures were performed in a conventional manner, and the EBUS images, pathology reports of the examined LNs, and clinical characteristics were all collected retrospectively.
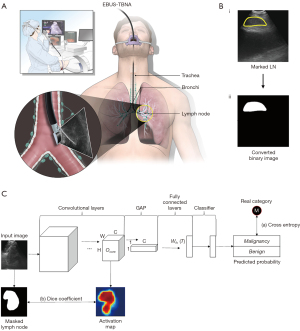
A total of 672 patients who underwent an EBUS-TBNA procedure were screened consecutively. Images that showed more than two LNs in one view, and Doppler overlaid images, were excluded (Figure S1).
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional review board at Yonsei University College of Medicine and Severance Hospital (IRB #4-2020-0857) and individual consent for this retrospective analysis was waived.
Malignant LN masking and cross-validation
Marking of the LNs was performed by the bronchoscopist-in-charge (SHY) without information on the pathology of the LNs to prevent bias in the selection of LN areas. The regions marked as LNs were converted into a binary image wherein each pixel was expressed as either 0 or 1 (Figure 1B). A 3-fold cross-validation was applied by using training, test, and validation sets. The training set was used to measure errors, which are back-propagated to the network to tune weight parameters. The validation set was used to check the training status by evaluating images at the end of each training epoch, while the evaluation set was used to assess the final performance after training completion. In the cross-validation fold, the validation and evaluation sets were each comprised of 200 images (100 malignant and 100 benign). The remaining images were used for the training set. The details of the 3-fold cross-validation are described in Table S1.
Network architecture and training
Network architecture, output format
To classify whether a LN that has been identified in the ultrasound image is malignant, the visual geometry group (VGG)-16 network (5) is employed. The architecture of the VGG-16 network used for this study is depicted in Figure S2. Because traditional CNNs, such as VGG-16, take the flattened output extracted from the output of the last convolutional layer as an input for the first fully connected layer, it is hard to operate CNNs in real-time on lite GPU devices. To operate the CNNs in real-time on these devices, we replaced the flattening operation with global average pooling (GAP) (6), requiring the matrix of the first fully connected layer that outputs the 4,096-dimensions to be of size 512×4,096. The GAP computes the global averages of the output from the last convolutional layer in a two-dimensional manner (width, height). Figure S2 shows the difference between the original VGG-16 and the VGG-16 with the GAP operation in its architecture. The last outputs of the classification model are two probability scores ranging from 0 to 1, and the summation of the two probabilities is 1.
Joint loss
To optimize the classification model during the training time, the gap between the predicted category (or probabilities) and the real category of input data was measured as a loss function. Because most of the distinguishable features between malignant and benign are located in the LN regions of the input images, using only a general loss function (e.g., cross-entropy loss) to train the model can lead to dataset bias, which results in the model locating local features outside of the LN regions. To guarantee the generalized classification performances of the model, we designed a new loss function that allows the model to focus on LN regions by providing local guidance for the LN regions, while also measuring the classification loss. By multiplying the output of the last convolutional layer and the operation matrix of the last fully connected layer, we can obtain the class activation map (CAM) (7). Each pixel of the CAM image has a value between 0 to 1, and pixels with higher values indicate a relatively higher contribution to the final decision of malignancy. We then computed the local differences between the activated map and the masked LN regions, using the dice coefficient (8). Subsequently, the dice coefficient value was added to the classification loss measured by the cross-entropy function. Figure 1C depicts an overview of the proposed loss function. Because the model, which was trained using the proposed loss function, considers the spatial information of LNs to predict the malignancy more precisely, the boundaries of the CAM output can be close to the actual LN regions. The boundaries extracted from the CAM output refer to the regions where pixels had a higher contribution to the final classification outputs of the trained network than the rest of the image regions.
Implementation setup
To build the proposed model, TensorFlow (version 2.4) installed on Python (version 3.7.10) was used. SciPy (version 1.7) and Matplotlib (version 3.4.2) were also utilized to compute the performance scores of deep learning. All setups were implemented on an NVIDIA GeForce RTX 2080Ti with 11 GB of memory for CUDA computations, whereas an NVIDIA GeForce RTX 2060 with 6 GB of memory was used to measure the execution times.
Statistical analyses
To clarify the generalized classification performance, the threshold was set to 0.5. In other words, LN images with a probability of more than 0.5 were decided as a malignancy. If the threshold value was selected according to the effective range of classification performance on our test dataset, it would be hard to claim that the performance represents the generalized result on any input data. To evaluate the performance of the malignancy prediction model, we used the sensitivity (%), specificity (%), accuracy (%), the area under the curves (AUC) by summing all cross-validation folds, and the average execution time for classifying an input image using pathologic reports as the gold standard.
To analyze the effectiveness of the automatically localized LN from the CAM output, we tracked the performance changes by thresholding the overlap ratio to the LN regions tagged by the bronchoscopist. The intersection over union (IoU) was used to measure the overlap ratio between the LN regions tagged by the bronchoscopist () and automatically localized regions () as follows:
where and are a binary image format. All summaries and analyses were performed using SPSS (version 26).
Results
Demographic and clinical characteristics
A total of 2,394 ultrasound images, including 1,459 benign LN images from 193 patients and 935 malignant LN images from 177 patients, were analyzed. All LN images were generated from 888 LNs, all of which were histologically confirmed. Among the 310 patients, 60 patients had both benign and malignant LNs. The demographics of patients and their clinical characteristics are presented in Table 1. The median age of patients was 67.0 years old, and there were more male patients than females (68.7% male). Of all diagnosed diseases, malignancies comprised 267 cases (86.1%), whereas benign diseases comprised 43 cases (13.9%). Among the malignancies, primary lung adenocarcinoma was the most common (61.4%), followed by lung squamous cell carcinoma (17.2%), small cell lung cancer (12.4%), and metastases from other organs (4.9%). All patients underwent chest computed tomography (CT) scans before EBUS-TBNA. The histology of 888 LNs was confirmed using EBUS-TBNA, and/or surgical resection performed immediately after EBUS-TBNA. Among all confirmed LNs, malignant LNs comprised 340 (38.3%), and benign LNs comprised 548 (61.7%). The most frequent sampling site was a subcarinal LN (26.4%), followed by a right lower paratracheal LN (20.9%). The median number of transbronchial needle aspiration (TBNA) for each LN was 2.0 (interquartile range, 1.0–2.0).
Table 1
| Patients or LNs characteristics | Value |
|---|---|
| Age, years old, median ± SD | 67.0±11.4 |
| Sex, n (%) | |
| Male | 213 (68.7) |
| Female | 97 (31.3) |
| Patients, total n | 310 |
| Who had malignant LNs | 177 |
| Who had benign LNs | 193 |
| Who had both malignant and benign LNs | 60 |
| Patients tumor histology, n (%) | |
| Malignant diseases | 267 (86.1) |
| Lung adenocarcinoma | 164 (61.4) |
| Lung squamous cell carcinoma | 46 (17.2) |
| Small cell lung cancer | 33 (12.4) |
| Other primary lung cancer | 11 (4.1) |
| Metastatic malignancy | 13 (4.9) |
| Benign diseases | 43 (13.9) |
| Histologic confirmed LNs, total n | 888 |
| Lymph nodes histology, n (%) | |
| Malignant | 340 (38.3) |
| Benign | 548 (61.7) |
| Lymph nodes station, n (%) | |
| Subcarinal | 234 (26.4) |
| Right lower paratracheal | 186 (20.9) |
| Left lower paratracheal | 83 (9.3) |
| Right hilar | 60 (6.8) |
| Left hilar | 58 (6.5) |
| Right interlobar | 100 (11.3) |
| Left interlobar | 145 (16.3) |
| Other (right upper paratracheal, left upper paratracheal, para-aortic, paraesophageal, lobar) | 22 (2.5) |
LNs, lymph nodes; SD, standard deviation.
Malignancy prediction performance using the modified VGG-16
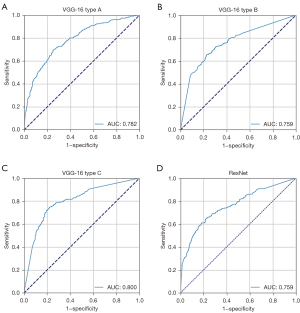
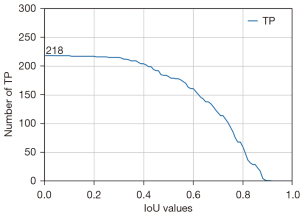
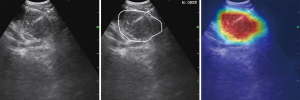
We tested and modified different VGG networks and evaluated their malignancy prediction performance (Figure 2A-2D and Table S2). First, we evaluated the VGG-16 with its original architecture, trained using only traditional cross-entropy for the classification loss (VGG-16 type A). The sensitivity, specificity, accuracy, and AUC of the malignancy prediction were 69.7%, 74.3%, 72.0%, and 0.782, respectively (Figure 2A). VGG-16 type A’s total time for predicting malignancy from an image was 0.021 seconds. We then evaluated the VGG-16 in which the flattening operation was replaced with a GAP operation, which was trained by using the classification loss (VGG-16 type B). The sensitivity, specificity, and accuracy for predicting malignancy were 68.3%, 72.7%, and 70.5%, respectively, and the overall AUC was 0.759 (Figure 2B). VGG-16 type B’s total time to process a single input image was, on average, 0.0159 seconds. In other words, approximately 63 images could be processed per second. In addition, we trained a residual network (ResNet), which was used as a backbone network by Lin et al. (4). The time taken for ResNet to predict the malignancy of an EBUS image was 0.0427 seconds. The sensitivity, specificity, accuracy, and AUC were 64.7%, 77.7%, 71.2%, and 0.759, respectively (Figure 2D). Finally, we examined the performance of the VGG-16 when the new loss function was applied to train the network (VGG-16 type C). Using this modified VGG-16, the overall AUC improved to 0.800 (Figure 2C). The sensitivity, specificity, and accuracy improved to 72.7%, 79.0%, and 75.8%, respectively. Figure 3 shows the changes in the number of true positives (TPs) according to the IoU thresholds. As shown in the figure, the number of TPs was stable until the IoU threshold reached 0.4. The representative image demonstrating the performance of the VGG-16 type C is shown in Figure 4 and Video S1.
Comparison with malignancy prediction by sonographic feature classification
We analyzed the sonographic features described by an experienced bronchoscopist at the time of the EBUS-TBNA procedure on an electronic medical record. Sonographic features were described based on a standard EBUS image classification system suggested by Fujiwara et al. (9): greater than 1 cm in size, round shape, distinct margin, heterogeneous echogenicity, absence of central hilar structure, and presence of coagulation necrosis sign indicated a high possibility of malignancy. For malignancy prediction performance, our data showed that shape had the highest accuracy (85.1%, P<0.001), followed by echogenicity (71.3%, P<0.001; Table 2). In the multivariate analysis, margin showed the highest odds ratio (OR) =31.1 [P<0.001; 95% confidence interval (CI): 5.0–195.0], followed by shape (OR =28.3; P<0.001; 95% CI: 10.5–76.2; Table S3).
Table 2
| Characteristic | Sensitivity (%) | Specificity (%) | NPV (%) | PPV (%) | Accuracy (%) | P value |
|---|---|---|---|---|---|---|
| Shape | 83.6 | 86.1 | 88.6 | 80.3 | 85.1 | <0.001 |
| Margin | 28.7 | 97.3 | 93.9 | 48.0 | 56.4 | <0.003 |
| Echogenicity | 76.7 | 67.6 | 81.1 | 61.5 | 71.3 | <0.001 |
| Central hilar structure | 72.6 | 31.5 | 63.0 | 41.7 | 48.1 | 0.642 |
| Coagulation necrosis sign | 13.7 | 97.2 | 62.5 | 76.9 | 63.5 | 0.213 |
NPV, negative predictive value; PPV, positive predictive value.
Discussion
In this study, we built a modified VGG-16 network to classify malignant thoracic LNs and support the selection of LNs for biopsy during EBUS-TBNA. EBUS-TBNA is a minimally invasive procedure, conducted under adequate sedation using a combination of fentanyl and midazolam (moderate sedation), propofol or general anesthesia (deep sedation). Moderate sedation is preferred for patient safety, but the diagnostic yields tend to be worse under moderate sedation than deep sedation (46.1–85.7% vs. 52.3–100.0%) (10). Owing to the limitation of sedation time, moderate sedation may not be sufficient for comprehensive staging or small LN sampling, especially for inexperienced bronchoscopists. Current guidelines for lung cancer staging mandate mediastinal staging, with a minimum of five LNs assessed by EBUS, and three LNs sampled systematically. However, this recommendation is completed in less than 50% of patients (11). This study was designed to build a deep learning model to support the decision-making of bronchoscopists for sufficient mediastinal assessment in shorter procedure times.
Until now, many efforts have been made to identify potential malignant LNs that require biopsy based on the size of the LNs, measured via CT scans, and the standardized uptake value (SUV), measured through fluorodeoxyglucose-positron emission tomography (FDG-PET) scans. However, it has been established that the LN size on a CT scan is not a good predictor of nodal metastasis in lung cancer (12). FDG-PET scans for mediastinal staging have limitations of both high false positive (FP) and false negative (FN) rates (13). In particular, micrometastases might occur within LNs in lung adenocarcinoma (14), so it is inaccurate to predict metastasis only by the size of the LNs on a CT scan or by the SUV on the FDG-PET scan. Therefore, sonographic features must be observed during EBUS-TBNA to select LNs for biopsy. However, various ultrasonographic features have different predictive values for malignancy depending on the observer. The feature that is identified as the strongest sonographic predictor differs depending on reports. Evison et al. (12) reported echogenicity and Wang Memoli et al. (15) reported shape as the strongest feature. In our data, shape showed the highest accuracy, followed by echogenicity. However, there is a limit to predicting malignancy based on each specific characteristic of the ultrasound. Therefore, it is possible to improve the predictive value of malignancy when comprehensively analyzing various characteristics on the ultrasound simultaneously. For this reason, Hylton et al. (16) developed the four-point scoring system using four sonographic features: short-axis diameter, margins, central hilar structure, and necrosis, and showed good performance in identifying malignant LNs. If all of these sonographic features can be evaluated comprehensively and simultaneously using a deep learning model, the model will be a robust predictor for the classification of malignant LNs.
Medical image analysis using a deep learning model has advantages, as it can process huge amounts of information rapidly and identify features at a microscopic level that cannot be confirmed visually by humans. Although deep learning has been widely applied to ultrasound images of other organs, there are few studies on the application of deep learning to EBUS images. Ozcelik et al. (17) recently developed an artificial neural network guided analysis and applied it to 345 LN images obtained during EBUS. They reported a diagnostic accuracy of 82% and AUC of 0.78. Similarly, Li et al. (18) applied deep learning to EBUS images of 294 LNs, and reported a diagnostic accuracy of 88%, with AUC of 0.95. These studies have reported good diagnostic accuracy with acceptable sensitivity and specificity. However, by analyzing only static images, they had the limitation of not applying their method directly to the dynamic images, which we achieve in our real-time EBUS procedure. Lin et al. (4) reported a three-dimensional CNN with ResNet as a backbone that uses video data directly and is robust against data noise. However, the size of the backbone network is large, and since more than one model is used, a real-time operation may be difficult on mainstream hardware. In our test, the ResNet could process approximately 23 EBUS images in a second. Considering that most ultrasound processors capture 30 or more frames per second, it is hard to run the ResNet architecture in real-time. To the best of our knowledge, we are the first to report the use of deep convolutional networks on EBUS images that can run as an application in real-time in current healthcare settings, without additional high-end hardware.
The strength of our model is that the proposed network can be applied on a mainstream GPU device during real-time EBUS-TBNA procedures. When the flattening operation was replaced with the GAP operation, the sensitivity only decreased by approximately 1.3%. However, the required processing time for a single input image was reduced by about 0.005 seconds, consequently increasing the number of processable images per second from 47 to 62. In other words, there were only a few performance decreases, while the complexity of the model was significantly reduced. Additionally, the proposed model achieved improved sensitivity (↑4.4%), specificity (↑6.4%), accuracy (↑5.3%), and AUC (↑0.041) over the model trained using the traditional cross-entropy for malignancy prediction performance.
This study has a limitation because it was designed as a retrospective study and developed based on EBUS image data from a single institution. To enhance the clinical usefulness of this deep learning model, external validation through multicenter prospective randomized trials is needed.
In conclusion, deep CNNs have shown effectiveness in classifying malignant LNs on EBUS images, with high accuracy. Deep learning can shorten the EBUS-TBNA procedure time, increase the accuracy of nodal staging, and improve patient safety. Therefore, evaluation of the real-world clinical benefit of this model in prospective trials is warranted.
Acknowledgments
The authors thank Medical Illustration & Design, part of the Medical Research Support Services of Yonsei University College of Medicine, for all artistic support related to this work.
Funding: This study was supported by a faculty research grant from Yonsei University College of Medicine (6-2021-0034) and Waycen Inc.
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-21-870/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-21-870/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-21-870/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-21-870/coif). SIO and JSK have been full-time employees of Waycen Inc. during this study. KNK has been the CEO of Waycen Inc. during this study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional review board at Yonsei University College of Medicine and Severance Hospital (IRB #4-2020-0857) and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Silvestri GA, Gonzalez AV, Jantz MA, et al. Methods for staging non-small cell lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013;143:e211S-50S.
- Hylton DA, Turner J, Shargall Y, et al. Ultrasonographic characteristics of lymph nodes as predictors of malignancy during endobronchial ultrasound (EBUS): A systematic review. Lung Cancer 2018;126:97-105. [Crossref] [PubMed]
- van Sloun RJ, Cohen R, Eldar YC. Deep learning in ultrasound imaging. Proceedings of the IEEE 2019;108:11-29. [Crossref] [PubMed]
- Lin K, Wu H, Chang J, et al. The interpretation of endobronchial ultrasound image using 3D convolutional neural network for differentiating malignant and benign mediastinal lesions. arXiv 2021. arXiv:210713820.
- Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv 2014. arXiv:14091556.
- Lin M, Chen Q, Yan S. Network in network. arXiv 2013. arXiv:13124400.
- Zhou B, Khosla A, Lapedriza A, et al. Learning deep features for discriminative localization. In: Proceedings of the IEEE conference on computer vision and pattern recognition. Las Vegas, NV, USA: IEEE, 2016.
- Sorensen TA. A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. Biologiske Skrifter 1948;5:1-34.
- Fujiwara T, Yasufuku K, Nakajima T, et al. The utility of sonographic features during endobronchial ultrasound-guided transbronchial needle aspiration for lymph node staging in patients with lung cancer: a standard endobronchial ultrasound image classification system. Chest 2010;138:641-7. [Crossref] [PubMed]
- Aswanetmanee P, Limsuwat C, Kabach M, et al. The role of sedation in endobronchial ultrasound-guided transbronchial needle aspiration: Systematic review. Endosc Ultrasound 2016;5:300-6. [Crossref] [PubMed]
- Hylton DA, Kidane B, Spicer J, et al. Endobronchial Ultrasound Staging of Operable Non-small Cell Lung Cancer: Do Triple-Normal Lymph Nodes Require Routine Biopsy? Chest 2021;159:2470-6. [Crossref] [PubMed]
- Evison M, Morris J, Martin J, et al. Nodal staging in lung cancer: a risk stratification model for lymph nodes classified as negative by EBUS-TBNA. J Thorac Oncol 2015;10:126-33. [Crossref] [PubMed]
- Detterbeck FC, Falen S, Rivera MP, et al. Seeking a home for a PET, part 2: Defining the appropriate place for positron emission tomography imaging in the staging of patients with suspected lung cancer. Chest 2004;125:2300-8. [Crossref] [PubMed]
- Carretta A. Clinical value of nodal micrometastases in patients with non-small cell lung cancer: time for reconsideration? J Thorac Dis 2016;8:E1755-8. [Crossref] [PubMed]
- Wang Memoli JS, El-Bayoumi E, Pastis NJ, et al. Using endobronchial ultrasound features to predict lymph node metastasis in patients with lung cancer. Chest 2011;140:1550-6. [Crossref] [PubMed]
- Hylton DA, Turner S, Kidane B, et al. The Canada Lymph Node Score for prediction of malignancy in mediastinal lymph nodes during endobronchial ultrasound. J Thorac Cardiovasc Surg 2020;159:2499-2507.e3. [Crossref] [PubMed]
- Ozcelik N, Ozcelik AE, Bulbul Y, et al. Can artificial intelligence distinguish between malignant and benign mediastinal lymph nodes using sonographic features on EBUS images? Curr Med Res Opin 2020;36:2019-24. [Crossref] [PubMed]
- Li J, Zhi X, Chen J, et al. Deep learning with convex probe endobronchial ultrasound multimodal imaging: A validated tool for automated intrathoracic lymph nodes diagnosis. Endosc Ultrasound 2021;10:361-71. [Crossref] [PubMed]


