
A multicenter-retrospective study of non-small-cell lung carcinoma harboring uncommon epidermal growth factor receptor (EGFR) mutations: different subtypes of EGFR exon 19 deletion-insertions exhibit the clinical characteristics and prognosis of non-small cell lung carcinoma
Introduction
Over the last few decades, lung cancer has become the leading cause of death from cancer worldwide. According to global cancer statistics from 2020, 2.2 million new cases were identified, which contributed 11.4% to the total cases and 1.8 million patients died of lung cancer globally, which comprised 18% of the total cancer deaths (1). Non-small cell lung cancer (NSCLC) accounts for 80–85% of lung cancer cases (2). Epidermal growth factor receptor (EGFR) gene mutations have become the predominant type, representing 10–20% and 40–50% of NSCLC cases in Caucasians and East Asians, respectively (3). The most frequent EGFR mutation subtype is exon 19 deletion (19del), involved in approximately 50% of all EGFR mutations (4). The large, randomized phase III randomized controlled trials (RCTs) showed that the objective response rate was 71.2% with gefitinib versus 47.3% with chemotherapy (5). Consequently, targeted therapy was approved by the Food and Drug Administration (FDA) for the first-line treatment of the patients with NSCLC with sensitive EGFR mutations (exon 19 deletion or L858R point mutation). In these double-blind 3 trials, the first-, second- or third-generation tyrosine kinase inhibitors (TKIs) currently represent the median progression-free survival (mPFS) of 9–13, 12–16, 21.4 m for NSCLC patients harboring sensitizing EGFR exon 19 deletion mutations (5-8).
It is well known that EGFR exon 19 deletion mutant tumors are sensitive to TKI; however, a large number of variants with different starting locations and with or without amino acid insertion/substitution have not been distinguished. There has been little detailed exploration of potential differences in TKI sensitivity among exon 19 deletion subtypes, especially the complex alteration of deletion-insertions. When we searched the literature, we did not a series of reports about the EGFR exon 19 deletion-insertions. Therefore, we performed a retrospective analysis to assess the clinical characteristics, immunohistochemistry (IHC), compound mutations sensitivity to different generation TKIs, and prognosis of the rare mutations of exon 19 deletion-insertions using next-generation sequencing (NGS). We present the following article in accordance with the STROBE reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-48/rc).
Methods
Patients
This retrospective analysis included 4,666 NSCLC patients harboring EGFR mutations in a multi-center study from January 2017 to December 2020 (including The 900th Hospital of the Joint Logistic Support Force, PLA; Fujian Provincial Hospital; Fujian Medical University Union Hospital; Zhangzhou Hospital Affiliated to Fujian Medical University; The Second Affiliated Hospital of Fujian Medical University; The First Affiliated Hospital of Xiamen University and Zhongshan Hospital Affiliated to Xiamen University). All patients who met the following criteria were included in the analysis: confirmed lung adenocarcinoma by pathology; NGS was used to detect the subtype of EGFR exon 19 deletion-insertions; responses were measured according to the Response Evaluation Criteria in Solid Tumors version 1.1 (RECIST 1.1); and complete clinical and first-line treatment data. Patients were excluded if they had a second primary malignant tumor and a concomitant mutation in 2 or more EGFR exons. In addition, patients whose diagnosis was made after December 2019 were also excluded to ensure a minimum follow-up period of 1 year. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of the 900th Hospital of the Joint Logistic Support Force, PLA (No. 2021-026) and all hospitals where the included patients are from have approved the study. Informed consent was taken from all the patients.
Data collection and treatment efficacy assessment
Clinical variables including age, gender, smoking, gene mutation, histologic type, IHC, pathological stage, the first line of therapy, and best response were collected by reviewing the medical charts and pathology records. The imaging data mainly including computed tomography (CT) scans and magnetic resonance imaging (MRI) scans, which were independently reviewed by the authors to evaluate their treatment responses according to RECIST 1.1. Cross-checking was performed for response evaluation to mitigate the problem of measurement errors. Types of response included complete response (CR), partial response (PR), stable disease (SD), or progressive disease (PD). The best response of each patient was recorded. Progression-free survival (PFS) or disease-free survival (DFS) was calculated from the date of the initial treatment to a radiologic or clinical observation of the disease progression. Patients were followed up from pathology diagnosis until progression of disease.
NGS detection
We obtained samples of blood or tumor tissue for biomarker analysis and transferred them to a designated central laboratory for NGS detection (Amoy Diagnostics, Xiamen, China). We conducted deoxyribonucleic acid (DNA) sequencing using a targeted 448-gene panel or a targeted 10-gene panel with a Next Seq 500 (Illumina Inc., San Diego, CA, USA) at Amoy Diagnostics Co., Ltd. Briefly, genomic DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tumor tissue or a blood sample, then processed through fragmentation, end-repairing, ligation of adapters, library amplification, and hybridization capturing. In order to ensure adequacy of sequences and mutation detection, at least 20% tumor area on each slice was set as a minimum. The kit covered the targeted drug-related hot spot mutation regions of 7,007 critical exons in 448 genes, and 59 introns in 26 fusion genes and promoter. The ultra-deep coverage of genes of interest was 2,000× for tumor tissue and 5,000× for serum. The sequence data were analyzed using the Amoy Dx NGS Data Analysis System ADXLC10 module (Amoy Diagnostics Co., Ltd.). An OncoPrint plot of circulating tumor DNA (ctDNA) profiled from 69 patients was generated for the most frequently mutated in NSCLC in The Cancer Genome Atlas (TCGA) dataset and included in the targeted gene panel.
Statistical analysis
Statistical analyses were performed with SPSS version 26.0 (IBM Corp., Armonk, NY, USA). Chi-square test and Fisher’s exact test were conducted to analyze the correlations between EGFR exon 19 deletion-insertion and clinicopathological variables. For the subgroup analysis, the same method was used to calculate PFS, hazard ratio (HR), and 95% confidence interval (CI) after categorizing the participants by age, gender, smoking status, stage, EGFR mutation status, the first line of the therapy, and best response. To perform a sensitivity analysis, we defined the subgroup of EGFR exon 19 deletion-insertion such as L747_P753delinsS, L747_A750delinsP, L747_T751delinsP, and the others. We drew PFS curves using the Kaplan-Meier method, and statistical differences were calculated by a 2-sided log-rank test. The Cox regression model was introduced to identify the predictive effect on PFS of different variables. A 2-sided P<0.05 was considered statistically significant. GraphPad Prism (GraphPad Inc., La Jolla, CA, USA) was used to generate survival curves and a complex heatmap was constructed (Figure 1).
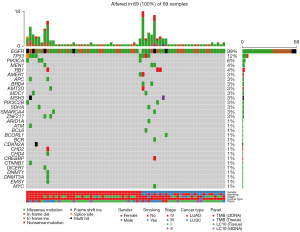
Results
Clinical characteristics of patients with EGFR exon 19 deletion-insertion
A total of 4,666 cases of primary lung adenocarcinomas underwent NGS testing, including 172 cases (3.7%) of EGFR exon 18 mutation, 959 cases (20.5%) of EGFR exon 19 mutation, 519 cases (11.1%) of EGFR exon 20 mutation, and 917 cases (19.5%) of EGFR exon 21 mutation (Figure 2). Based on selection criteria, a total of 69 patients with EGFR exon 19 deletion-insertion were taken to account in the study, which comprised 24 cases (34.8%) in L747_P753delinsS, 9 cases (13.1%) in L747_A750delinsP, both 5 cases (7.2%) in E746_A750delinsQP, E746_S752delinsV and both 4 cases (5.8%) in E746_T751delinsA and L747_T751delinsP (Figure 3). Of 69 patients with baseline NGS data, all had EGFR exon 19 deletion-insertion mutations and were concurrent with tumor-suppressor gene mutations. The OncoGrid depicts gender, smoking, stage, cancer-type, and panel with the subtypes of EGFR exon 19 deletion-insertion in lung cancer. The OncoGrid showed frequencies including a composite of all alterations (In_Frame_Del, In_Frame_Del, Frame_Shift_Ins, Frame_Shift_Del, and Silent). Gene mutation status analyzed by NGS revealed that compound mutations were recombinant tumor protein 53 (TP53) (45.83%), phosphatidylinositol 3-kinases (PIK3CA) (11.59%), and almost 16.67% in retinoblastoma 1 (RB1), melanocyte stimulating hormone (MSH), and myelocytomatosis (MYC) (Table 1, Figure 1). Among the samples, there were 29 males (42%) and 40 females (58%), with a median age of 59.7 years (range, 29 to 88 years). Of those, 12 (21.7%) participants were smokers and 54 (78.3%) were nonsmokers. As shown in Table 1, all participants were divided into stage I (43.5%), stage II (1.5%), stage III (5.8%), and stage IV (49.2%) according to the tumor-node-metastasis (TNM) staging system. They received different treatments according to stage of disease, including surgery (47.8%), chemotherapy (1.5%) and targeted therapy (50.7%). The best response for the first line of the therapy exhibited SD (13.1%), PR (33.3%), and CR (47.8%). The sum of stable, partial, and the disease control rate (DCR) was 29% of patients at 10 months follow-up (Figure 4). The characteristics of these patients are listed in Figures 1-4, and Table 1.
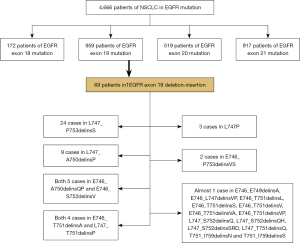
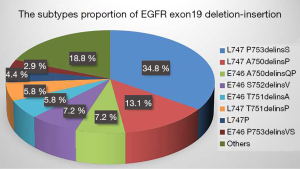
Table 1
| Clinical characteristics | N | % |
|---|---|---|
| Median age [range], years | 59.7 [29–88] | |
| Gender | ||
| Male | 29 | 42.0 |
| Female | 40 | 58.0 |
| Smoking | ||
| Yes | 15 | 21.7 |
| No | 54 | 78.3 |
| Compound mutation | ||
| TP53 | 11/24 | 45.83 |
| PIK3C | 8/69 | 11.59 |
| RB1 | 4/24 | 16.67 |
| MSH | 4/24 | 16.67 |
| MYC | 4/24 | 16.67 |
| Unknown | 45 | 65.22 |
| Immunohistochemistry | ||
| TTF-1 | 53 | 93.0 |
| Napsin A | 52 | 89.7 |
| CK7 | 31 | 98.1 |
| B-tubulin | 8 | 100 |
| SPB | 10 | 83.3 |
| CD56 | 3 | 18.5 |
| TNM stage | ||
| I | 30 | 43.5 |
| II | 1 | 1.5 |
| III | 4 | 5.8 |
| IV | 34 | 49.2 |
| The first line of the therapy | ||
| Surgery | 33 | 47.8 |
| Targeted therapy | 35 | 50.7 |
| Chemotherapy | 1 | 1.5 |
| Best response | ||
| CR | 33 | 47.8 |
| PR | 23 | 33.3 |
| SD | 9 | 13.1 |
| PD | 4 | 5.8 |
TP53, tumor protein 53; PIK3C, phosphoinositide-3-kinase; RB1, retinoblastoma 1; MSH, melanocyte stimulating hormone; MYC, myelocytomatosis; TTF, thyroid transcription factor; CK7, cytokeratin; SPB, surfactant proteins B; TNM, tumor-node-metastasis; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease.
Correlation between immunoreactivity of Napsin A, thyroid transcription factor (TTF), cytokeratin (CK7), surfactant proteins B (SPB), and the subtypes of EGFR exon 19 deletion-insertion
The results of IHC for each subtype of EGFR exon 19 deletion-insertion of adenocarcinomas are summarized in Table 2. The chi-square test and Fisher’s exact test were conducted to analyze the correlations between EGFR exon 19 deletion-insertion and IHC markers. Based on the analysis, we found there was significantly statistical difference of IHC expression among different types of EGFR exon 19 deletion-insertion. Napsin A and TTF had higher expression ratios on these subtypes of EGFR exon 19 deletion-insertion, ranging from 66.7% to 100% (median: 93.5%) and 90% to 100% (median: 97.8%). Compared with CK7 and SPB, they had different expression ratios. It did not show the expression ratios of CK7 on E746_P753delinsVS. The expression ratios of SPB were 0 on L747_T751delinsP and L747_T751delinsP.
Table 2
| Exon 19 deletion-insertion subtypes | Immunohistochemistry (%) | ||||
|---|---|---|---|---|---|
| Napsin A | TTF-1 | CK7 | SPB | P value | |
| L747_P753delinsS | 85 | 90 | 94 | 100 | <0.05 |
| L747_A750delinsP | 100 | 100 | 100 | 100 | |
| E746_A750delinsQP | 100 | 100 | 100 | 50 | |
| E746_S752delinsV | 100 | 100 | 100 | 100 | |
| E746_T751delinsA | 100 | 100 | 100 | 100 | |
| L747_T751delinsP | 100 | 100 | 100 | 0 | |
| L747P | 66.7 | 100 | 100 | 100 | |
| E746_P753delinsVS | 100 | 100 | 0 | 0 | |
| Others | 90 | 90 | 90 | 50 | |
EGFR, epidermal growth factor receptor; IHC, immunohistochemistry; TTF, thyroid transcription factor; CK7, cytokeratin; SPB, surfactant proteins B.
PFS according to clinical features and EGFR exon 19 deletion-insertion subtypes
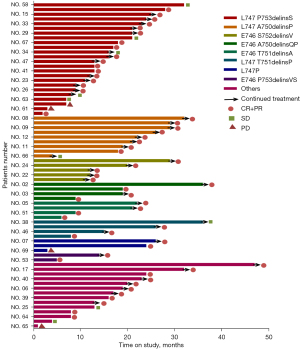
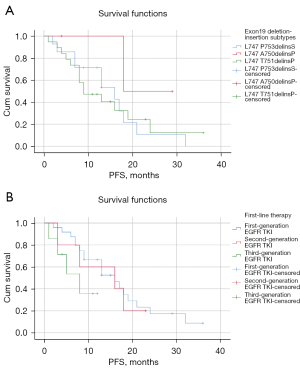
Either PFS or DFS was available for these patients. At the time of the data cut-off (31 May 2021), there were 36 patients with stage IV NSCLC in EGFR exon 19 deletion-insertion undergoing analysis. A total of 26 patients had experienced disease progression or death and the others were still undergoing follow-up. As shown in Table 3, the Kaplan-Meier method and Cox regression model included age, gender, smoking status, exon 19 deletion-insertion subtypes, TNM stage, the first line of the therapy, and best response. The median PFS and 95% CI of exon 19 deletion-insertion subtypes were 14.821 (9.917 to 19.726) months for L747_P753delinsS, 23.500 (15.877 to 31.123) months for L747_A750delinsP, 26.667 (11.731 to 41.603) months for L747_T751delinsP, and 11.713 (7.786 to 15.639) months for the others (Table 3). Using the Kaplan-Meier method, we detected the subtypes of L747_A750delinsP and L747_T751delinsP achieved a longer median PFS (median: 23.5, 95% CI: 15.877 to 31.123; median: 26.667, 95% CI: 11.730 to 41.603) than the L747_P753delinsS subtype (median: 14.821, 95% CI: 9.917 to 19.726) (Figure 5A). Furthermore, the Cox regression model was performed on the participants. The HR was comparable in L747_A750delinsP and L747_P753delinsS (HR: 0.306, 95% CI: 0.039 to 2.415). Similar results were obtained by L747_P753delinsS, and the HR of L747_T751delinsP was 0.266 (95% CI: 0.033 to 2.112). The clinical characteristics of patients analyzed by Kaplan-Meier method and Cox proportional hazards model indicated that age, gender, and smoking status did not affect the survival of EGFR exon 19 deletion-insertion (all P>0.05). Comparison of PFS among the 3 different EGFR TKIs revealed that patients receiving treatment with the 3rd generation TKI had the shortest PFS (median: 7.179, 95% CI: 3.969–10.388). Notably, the first EGFR-TKI had the longest mPFS (median: 16.653, 95% CI: 12.460 to 20.847) for the EGFR exon 19 deletion-insertion subtypes (Figure 5B).
Table 3
| Characteristics | Median PFS in months (95% CI) | P value | HR (95% CI) | P value |
|---|---|---|---|---|
| Age at diagnosis, years | ||||
| <65 | 15.497 (10.825–20.172) | 1 (ref) | ||
| ≥65 | 14.791 (9.187–20.395) | 0.723 | 1.148 (0.525–2.510) | 0.730 |
| Gender | ||||
| Female | 13.376 (9.660–17.093) | 1 (ref) | ||
| Male | 16.794 (11.040–22.548) | 0.418 | 1.377 (0.621–3.055) | 0.432 |
| Smoking status | ||||
| No | 14.299 (10.523–18.076) | 1 (ref) | ||
| Yes | 17.400 (9.504–25.296) | 0.381 | 0.684 (0.285–1.644) | 0.396 |
| Exon 19 deletion-insertion subtypes | ||||
| L747_P753delinsS | 14.821 (9.917–19.726) | 1 (ref) | ||
| L747_A750delinsP | 23.500 (15.877–31.123) | 0.306 (0.039–2.415) | 0.261 | |
| L747_T751delinsP | 26.667 (11.730–41.603) | 0.266 (0.033–2.112) | 0.210 | |
| Others | 11.713 (7.786–15.639) | 0.184 | 1.333 (0.587–3.029) | 0.492 |
| The first line of the therapy | ||||
| First EGFR-TKI | 16.653 (12.460–20.847) | 1 (ref) | ||
| Second EGFR-TKI | 13.600 (7.313–19.887) | 1.136 (0.378–3.412) | 0.820 | |
| Third EGFR-TKI | 7.179 (3.969–10.388) | 0.216 | 2.731 (0.833–8.953) | 0.097 |
| Best response | ||||
| CR | 19.000 (19.000–19.000) | 1 (ref) | ||
| PR | 15.855 (12.440–19.270) | 1.222 (0.157–9.519) | 0.848 | |
| SD | 17.206 (9.555–24.858) | 0.000 | 1.144 (0.135–9.688) | 0.902 |
PFS, progression-free survival; EGFR, epidermal growth factor receptor; NSCLC, non-small cell lung cancer; HR, hazard ratio; CI, confidence interval; TKI, tyrosine kinase inhibitor; CR, complete response; PR, partial response; SD, stable disease.
Discussion
The identification of activating EGFR mutations led to a paradigm shift in the management of this oncogene-addicted subgroup of NSCLC patient. The EGFR TKIs were shown to substantially improve survival time of patients with lung cancer, and became the standard first-line therapy for advanced NSCLC with EGFR exon 19 or 21 mutation according to the National Comprehensive Cancer Network (NCCN) guidelines (2) To our knowledge, EGFR exon 19 mutation has different variants, and discordant results have been reported for the various subtypes of EGFR exon 19 mutation (9,10). Literature is currently scarce about EGFR exon 19 deletion-insertion on account of its low frequency. Our study retrospectively analyzed 69 patients with EGFR exon 19 deletion-insertion from a cohort of 4,666 cases, which were classified into 21 subtypes.
In our study, we found that the most common EGFR exon 19 deletion-insertion was L747_P753delinsS (34.8%), followed by L747_A750delinsP (13.1%). The proportion of them was the same as reported by Truini et al. (10). Compound mutations are rarely encountered in daily practice, for which the detection rate ranges from 4 to 14% (11,12). Single driver mutation is being challenged for the development of NGS, which is capable of sequencing multiple genes at a time. Our study adopted NGS-based repeated deep sequencing and provided higher typical and atypical compound mutations. As previously reported, the mutational frequency of PIK3CA was 5% and relatively high in squamous histology (10.1%) (13). In addition, the overall rate of TP53 in NSCLC was approximately 40% (14). However, the majority of compound mutations in EGFR exon 19 deletion-insertion were composed of TP53 (45.83%), PIK3CA (11.59%), RB1 (16.67%), MYC (16.67%), and so on. Our data suggested that L747_P753delinsS was the most common subtype accompanied with TP53, PIK3CA, RB1, and MYC. These findings indicate that additional biomarkers may play an important role in predicting response to TKI therapy. Therefore, the median PFS of L747_P753delinsS was shorter than that of L747_A750delinsP and L747_T751delinsP.
Moreover, we evaluated the correlation between the subtypes of EGFR exon 19 deletion-insertion and IHC. Napsin A, TTF, CK7, and SPB are tissue-specific markers for well-differentiated lung adenocarcinomas (15). In our findings, the subtypes of EGFR exon 19 deletion-insertion were highly expressed the IHC markers, including Napsin A, TTF, and CK7. However, the expression of IHC had significantly statistic differences in the subtypes of EGFR exon 19 deletion-insertion (P<0.05). In the report by Taguchi et al., SPB was significantly associated with lung cancer risk (16). In our data, we detected the subtypes of L747_T751delinsP and E746_P753delinsVS were not the expression of SPB. The expression of CD56 is seen in hematological malignancies, small cell lung cancer, and ovarian cancer, which serves as a diagnostic biomarker to identify those of neuroendocrine origin (17). In a cohort study, the CD56 expression of T751_I759delinsS was 100%. A potential explanation for this may be that the subtype tends towards neuroendocrine differentiation.
Among the mutations, EGFR exon 19 deletion is the most sensitive to TKIs (18). To our knowledge, the series open-label, randomized controlled trials had implied that the 3rd generation TKI achieved a 21.4-month PFS of exon 19 deletion (8). Yet, we found that EGFR exon 19 deletion-insertion mutations exhibit limited sensitivity to the 3rd generation TKI, for which the median PFS was 7.1 months. Moreover, the EGFR exon 19 deletion-insertion mutations were observed to have longer median PFS for the 1st and 2nd generation TKI compared with the 3rd generation TKI, for which the median PFS were only 16.65 months for the 1st generation TKI and 13.60 months for the 2nd generation TKI. Importantly, EGFR exon 19 deletion-insertion mutations comprise a heterogeneous group of genetic aberrations, including various in-frame deletions, substitutions, and insertions (19). A few researchers have reported the differences in therapeutic sensitivity with exon 19 deletion subtypes, but their conclusions have been controversial. On the one hand, Xu et al. (4). reported that deletion location and type of variants affect therapeutic efficacy; on the other hand, Rossi et al. (20). reported that different exon 19 deletions are equally sensitive to therapy. However, there was still an absence of a large cohort analysis of exon 19 deletion subtypes reported in the literature. Similarly, Truini et al. demonstrated that the L747_A750delinsP mutation exhibited significantly worse outcomes than those with tumors harboring L747_P753delinsS mutations (10). In contrast to our finding, the subtype of L747_T751delinsP achieved longer PFS, which indicated a better prognosis and L747_P753delinsS had shorter PFS. These findings highlight the importance of mutation-specific EGFR-TKI selection and have shown the precise nature of deletions in therapeutic effect.
Several limitations must be noted in our study. First, this was a retrospective study that may have resulted in bias in the assessment of statistical analysis. Second, we did not investigate a more detailed classification because of the small sample size of patients. In addition, length of time bias caused by varying intervals of therapy could have led to the differences observed in PFS, but the long-term observations were not evaluated such as overall survival rate.
In conclusion, the subtypes of exon 19 deletion-insertion exhibit different IHC markers and clinical outcomes to EGFR TKIs. Our findings argued in favor of analyzing the correlation between immunoreactivity and the subtypes of EGFR exon 19 deletion-insertion. Most of the EGFR exon 19 deletion-insertion mutations exhibited limited sensitivity to 3rd generation TKI, except for the L747_T751delinsP that achieved longer PFS.
Acknowledgments
The authors appreciate the academic support from the AME Lung Cancer Collaborative Group.
Funding: This work was supported by grants from the Clinical Key Specialty Construction Project of Fujian Province (Nos. 2015-593 and 2017YZ0001-2), the 900th Hospital of the Joint Logistic Support Force of China: International Cooperative Research Program (Grant/Award Number: 2017L03), and Outstanding Youth Project in the 900th Hospital of the PLA Joint Logistics Support Force (No. 2021JQ07).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-48/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-48/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-48/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of the 900th Hospital of the Joint Logistic Support Force, PLA (Approval No. 2021-026) and informed consent was taken from all the patients. All hospitals where the included patients are from have approved the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Erratum: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2020;70:313. [Crossref] [PubMed]
- Ettinger DS, Wood DE, Aisner DL, et al. Non-Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2017;15:504-35. [Crossref] [PubMed]
- Ikemura S, Yasuda H, Matsumoto S, et al. Molecular dynamics simulation-guided drug sensitivity prediction for lung cancer with rare EGFR mutations. Proc Natl Acad Sci U S A 2019;116:10025-30. [Crossref] [PubMed]
- Xu H, Li W, Yang G, et al. Heterogeneous Response to First-Generation Tyrosine Kinase Inhibitors in Non-Small-Cell Lung Cancers with Different EGFR Exon 19 Mutations. Target Oncol 2020;15:357-64. [Crossref] [PubMed]
- Hosomi Y, Morita S, Sugawara S, et al. Gefitinib Alone Versus Gefitinib Plus Chemotherapy for Non-Small-Cell Lung Cancer With Mutated Epidermal Growth Factor Receptor: NEJ009 Study. J Clin Oncol 2020;38:115-23. [Crossref] [PubMed]
- Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol 2012;13:239-46. [Crossref] [PubMed]
- Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013;31:3327-34. [Crossref] [PubMed]
- Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:113-25. [Crossref] [PubMed]
- Su J, Zhong W, Zhang X, et al. Molecular characteristics and clinical outcomes of EGFR exon 19 indel subtypes to EGFR TKIs in NSCLC patients. Oncotarget 2017;8:111246-57. [Crossref] [PubMed]
- Truini A, Starrett JH, Stewart T, et al. The EGFR Exon 19 Mutant L747-A750>P Exhibits Distinct Sensitivity to Tyrosine Kinase Inhibitors in Lung Adenocarcinoma. Clin Cancer Res 2019;25:6382-91. [Crossref] [PubMed]
- Kobayashi S, Canepa HM, Bailey AS, et al. Compound EGFR mutations and response to EGFR tyrosine kinase inhibitors. J Thorac Oncol 2013;8:45-51. [Crossref] [PubMed]
- Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129-39. [Crossref] [PubMed]
- Sawa K, Koh Y, Kawaguchi T, et al. PIK3CA mutation as a distinctive genetic feature of non-small cell lung cancer with chronic obstructive pulmonary disease: A comprehensive mutational analysis from a multi-institutional cohort. Lung Cancer 2017;112:96-101. [Crossref] [PubMed]
- Bar J, Damianovich M, Hout Siloni G, et al. Genetic mutation screen in early non--small-cell lung cancer (NSCLC) specimens. Clin Lung Cancer 2014;15:159-65. [Crossref] [PubMed]
- Zhang R, Li Y, Nie X, et al. Prognostic implications of immunohistochemistry markers for EGFR-TKI therapy in Chinese patients with advanced lung adenocarcinoma harboring EGFR mutations. Onco Targets Ther 2016;9:355-66. [PubMed]
- Taguchi A, Hanash S, Rundle A, et al. Circulating pro-surfactant protein B as a risk biomarker for lung cancer. Cancer Epidemiol Biomarkers Prev 2013;22:1756-61. [Crossref] [PubMed]
- Shah MH, Lorigan P, O'Brien ME, et al. Phase I study of IMGN901, a CD56-targeting antibody-drug conjugate, in patients with CD56-positive solid tumors. Invest New Drugs 2016;34:290-9. [Crossref] [PubMed]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [Crossref] [PubMed]
- Sutiman N, Tan SW, Tan EH, et al. EGFR Mutation Subtypes Influence Survival Outcomes following First-Line Gefitinib Therapy in Advanced Asian NSCLC Patients. J Thorac Oncol 2017;12:529-38. [Crossref] [PubMed]
- Rossi S, Toschi L, Finocchiaro G, et al. Impact of Exon 19 Deletion Subtypes in EGFR-Mutant Metastatic Non-Small-Cell Lung Cancer Treated With First-Line Tyrosine Kinase Inhibitors. Clin Lung Cancer 2019;20:82-7. [Crossref] [PubMed]


