
Rapid genomic profiling of circulating tumor DNA in non-small cell lung cancer using Oncomine Precision Assay with Genexus™ integrated sequencer
Introduction
It is known that 69% of patients with advanced non-small cell lung cancer (NSCLC) harbored an actionable oncogenic driver alteration (1). Hence, molecular profiling of tumor tissues has become a routine clinical practice to identify actionable alterations in genes such as EGFR, ALK, ROS1, NTRK, MET and BRAF, whose information is applicable for the selection of molecular targeted therapies. The response rates were reported to be 50–80% for patients who received molecularly-guided targeted therapy and overall survival periods ranged between 18.0 months to about 5 years (2-5).
Although tissue biopsies remains a standard procedure for molecular profiling, approximately one fourth of lung carcinoma patients have no or insufficient volume of cancer tissue to be evaluated (6-9). Hence, there is a necessity to identify alternative approaches to overcome this limitation. Recent advances enabled comprehensive genomic profiling with liquid biopsy, a highly-sensitive method to detect circulating tumor DNA (ctDNA) that is shed from tumor cells and released into the blood circulation. Liquid biopsies are easily accessible through minimally invasive blood sampling that can be performed repeatedly to access dynamic and longitudinal changes of tumor mutations throughout cancer management (10). Evaluation of ctDNA also provide a more comprehensive genomic landscape as it reflects multiple tumor regions, and address intratumor and intertumoral heterogeneity that was difficult to be achieved with tissue biopsy (10).
In recent years, the advancement of next generation sequencing (NGS) allowed simultaneous screening of a wider range of targetable genetic mutations, gene amplification, or rearrangement of genes with one assay. Currently, the US Food and Drug Administration (FDA) has approved several broad targeted NGS panels as clinical diagnostic tests for genomic profiling in routine clinical usage. These include MSK-IMPACT, FoundationOne and Oncomine Dx Target test for tumor biopsy as well as Guardant360 and FoundationOne Liquid for liquid biopsy. However, the turnaround time (TAT) using NGS approaches can take up from 20 to 40 days which are longer than the routine single gene test that resulted in the delay of providing the precise treatment to the patients (11,12).
To improve the TAT significantly, Ion Torrent™ Genexus™ Integrated Sequencer (Genexus) was developed to automate all targeted NGS workflows from library construction using either nucleic acids of formalin-fixed paraffin-embedded (FFPE) tissues or cell-free total nucleic acids (cfTNA) of plasma to data analyses and delivers results within 24 h. The Oncomine Precision Assay (OPA) is an actionable alterations-focused panel that targeted 50 genes covering 2,769 unique variants including hotspot mutations, copy number variations (CNV) and gene fusions (Figure S1, available online: https://cdn.amegroups.cn/static/public/tlcr-21-981-1.xlsx). The targeted genomic alterations correspond to molecular targeted drugs with labels, those in guidelines as well as in global clinical trials. In addition to actionable alterations, 218 potential resistance-associated mutations across 22 genes are also included in this assay. The Genexus-OPA system requires as little as 10–20 ng of DNA from tumor tissues or cell-free DNA to assess a total of 1,773 mutations and indels, 981 fusions and 14 copy number variations. In this study, we performed liquid biopsy molecular profiling of 119 advanced NSCLC using Genexus™ Integrated Sequencer and Oncomine Precision Assay (Genexus-OPA). We evaluated the concordance rate, sensitivity and specificity of genomic alterations detected from liquid biopsy by comparing with standard-of-care (SOC) clinical diagnostic tests using tumor tissues. To our knowledge, this is the first report to assess the performance of Genexus-OPA in detecting genomic alterations using liquid biopsy in NSCLC. We present the following article in accordance with the MDAR reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-21-981/rc).
Methods
Patient cohort
The eligibility criteria of patient recruitment in this study are patients who are either newly diagnosed with advanced (stage III or IV) or recurrent from NSCLC without indication of surgery. A total of 119 patients were enrolled between August 2020 and June 2021 at the Cancer Institute Hospital, Japanese Foundation for Cancer Research after exclusion of patients with other concurrent cancers. Fourteen milliliters of peripheral blood samples in EDTA-2Na tubes (Terumo, Tokyo, Japan) were collected at diagnosis or before starting 1st line treatment. Clinical information was obtained to examine clinicopathological association with ctDNA detection. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study design is approved by the ethical committee in Japanese Foundation for Cancer Research (No. IRB-2020-1055) and all patients provided written informed consent to this study.
Collected peripheral blood was centrifuged at 2,000 g at 4 ℃ for 10 min to separate plasma within 0.5–2 h after the sample collection. Subsequently, the plasma samples were further centrifuged at 16,000 g at 4 ℃ for 10 min to remove cell debris. The plasma samples were stored at −80 ℃ until cfTNA extraction.
Extraction of cfTNA
cfTNA, which includes both DNA and RNA, was extracted using MagMAX Cell-Free Total Nucleic Acid Isolation Kit (Applied Biosystems, Austin, TX, USA) according to the manufacturer’s protocol. The amount of extracted cfTNA was measured using Qubit DNA HS Assay Kit (Life Technologies, Oregon, US) and the fragment sizes were assessed with High Sensitivity D5000 ScreenTape (Agilent, California, USA).
Next generation sequencing
The Ion Torrent™ Genexus™ Integrated Sequencer is a fully automated NGS system that integrates library preparation, template preparation, sequencing and data analysis. This automated sequencer delivers a sample-to-result workflow within 24 h. The OPA selected for this feasibility study allows simultaneous detection of somatic mutations, copy number variations and fusions genes for 50 cancer-related genes. Frequently-mutated genes in NSCLC such as TP53, EGFR, BRAF, KRAS, ALK, ROS1, NTRK, and RET were included in OPA. The OPA assay is an amplicon-based assay with unique molecular indexes (UMI) to minimize NGS errors.
Before sequencing with clinical samples, the performance of OPA with Genexus™ Integrated Sequencer was evaluated with various NGS standard control samples include two FFPE DNA controls [5% variant allele frequency (VAF) FFPE standard controls (TFS), FFPE Structural Multiplex Reference Standard (Horizon Diagnostic, HD789)], two FFPE RNA fusion control [FFPE NTRK Fusion RNA Reference Material (Seraseq®, 0710-1031), FFPE Tumor Fusion RNA Reference Material (Seraseq®, 0710-0496)] and two cell-free DNA reference standards [0.25MM Plasma TNA Control (TFS) and Multiplex I cfDNA Reference Standard Set (Horizon Diagnostic, HD780)].
The optimal input of solid tumor (FFPE DNA or RNA) is 10 ng (validated range: 5–15 ng) and liquid biopsy (cfTNA) is 20 ng (validated range 5–40 ng) for Genexus-OPA. After assigning an assay that specified the type of sample and sequencing run settings to the Genexus Integrative system, samples were diluted and loaded to the sample plate. About 10–20 ng of standard control samples (available online: https://cdn.amegroups.cn/static/public/tlcr-21-981-2.xlsx) and a median of 20 ng (range, 11–36 ng) of cfTNA isolated from plasma of 119 NSCLC cases were incorporated into Genexus-OPA for this study. For cfTNA NGS, a maximum of four samples were included to the GX5TM chip in a single run that expected to yield an output of 12–15 million reads per samples. Sequencing data was mapped and analyzed using on-instrument Genexus software. Detected alterations were annotated using Oncomine Knowledgebase Reporter Software (Oncomine Reporter 5.0).
Comparison of genomic profiling between tumor tissues and liquid biopsy
Genomic profiles of tumor tissues were performed using standard-of-care (SOC) clinical diagnostic tests. These SOC mutation analyses using tumor tissue included: (I) EGFR mutation screening with Cobas EGFR test, (II) ALK fusion assessment with ALK-IHC and (III) Oncomine Dx target test that covered 9 frequently mutated genes (about 9 major genes; EGFR, ALK, ROS1, BRAF, MET, KRAS, ERBB2, NTRK and RET) in NSCLC was performed in CLIA certificated commercial laboratory (LSI Medience). The genomic profiles of the tumor tissues were compared with genetic alterations detected from plasma cfTNA using the Genexus-OPA liquid biopsy assay. Liquid biopsy NGS results would not be compared if the samples did not submit for Oncomine Dx target test.
Statistical analysis
Relative molecular coverage (RMC) of each amplicon was calculated using the following formula to evaluate the amplification efficiency of individual amplicon included in the OPA panel. With a threshold at 0.5, indicated the coverage of the specific amplicon is 50% of molecular depth compares to the median molecular coverage of the sample.
Univariate analyses were performed using Fisher’s exact test. Mann-Whitey U test was applied to compare continuous data. All statistical analyses were performed using EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan), which is a graphical user interface for R (The R Foundation for Statistical Computing, Vienna, Austria). All reported P values were two-sided, and a P value of <0.05 was considered statistically significant. The RMC boxplots were generated using R program (version 4.0.3).
Results
Patient characteristics
A total of 119 patients were enrolled in this study. The summary of clinical characteristics is shown in Table 1. The median age of the patients was 70 years (range, 32–86 years); 43 (36.1%) were females; 31 (26.1%) had never smoked; 86 (72.3%) had adenocarcinoma and 73 (61.3%) were diagnosed as stage IV. Standard-of-care (SOC) tumor mutation analysis using their tumor tissues were performed with Cobas EGFR test for 111 (93.3%) cases, with ALK-IHC for 106 (89.1%) cases and with Oncomine Dx for 89 (74.8%) cases.
Table 1
| Clinical characteristics | N=119 |
|---|---|
| Age (years), median [range] | 70 [32–86] |
| Gender (female/male) | 43/76 |
| Smoking (never/smoker) | 31/88 |
| Pathological types (adenocarcinoma/squamous cell carcinoma/other*) | 86/25/8 |
| Stage (III/IV/recurrence) | 22/73/24 |
| EGFR Cobas performed (yes/no) | 111/8 |
| ALK-IHC performed (yes/no) | 106/13 |
| Oncomine Dx performed (yes/no) | 89/30 |
*, other include non-small cell lung carcinoma, not otherwise specified and large cell neuroendocrine carcinoma. IHC, immunohistochemistry.
NGS analysis with Genexus-OPA
Sequencing results of NGS standard control samples with Genexus-OPA were summarized in website: https://cdn.amegroups.cn/static/public/tlcr-21-981-2.xlsx. The first batch of FFPE tissue DNA controls contained 74 single nucleotide variants (SNVs) and INDELs with median allele frequencies of 5%. These FFPE DNA controls were analyzed in duplicate runs for the evaluation of Genexus-OPA. The duplicated analysis of this control samples showed the median of variant allele frequencies of 4.6% (range, 2.6–9.3%) and 4.5% (range, 2.8–9.4%), respectively. The second FFPE DNA control set (FFPE Structural Multiplex Reference Standard) consisted of 15 alterations with allele frequencies ranged between 3–15%. The results of the duplicate runs are highly concordant with R2 value of 0.938. All the driver fusion genes in the two FFPE RNA controls (FFPE NTRK Fusion RNA Reference Material, FFPE Tumor Fusion RNA Reference Material) were detected in the frequencies ranged between 5–20% by the Genexus-OPA system. For plasma cfTNA standard controls [0.25MM Plasma TNA Control (TFS) and Multiplex I cfDNA Reference Standard Set (Horizon Diagnostic, HD780)], Genexus-OPA detected alterations with variant allele frequency ranging 0.09–0.91% with a median of 0.27% (available online: https://cdn.amegroups.cn/static/public/tlcr-21-981-2.xlsx).
Among the 119 NSCLC patients enrolled, a median of 50 ng (range, 14.7–1,521.3 ng) of cfTNA were extracted from the plasma samples separated from 14 mL of blood (a median volume of the plasma samples in these patients of 5.0 mL with a range of 2.5–7.1 mL) (Figure S2). 96% of samples achieved the optimal cfTNA input at 20 ng or more extracted from plasma (derived from 14 mL of blood collections) for Genexus-OPA sequencing system.
The detailed summary of NGS analysis parameters and the amounts of cfTNA input are shown in Table 2 and in website: https://cdn.amegroups.cn/static/public/tlcr-21-981-3.xlsx, respectively. In brief, a median reads per sample were 10,467,880 (range, 4,765,877–15,510,927). The median overall depth and median molecular coverage with median input of 20 ng (a range of 11.1–36.4 ng) were 35,773× (range, 13,485–55,882×) and 2,192× (a range of 590–3,311×), respectively. The median reads per functional molecules with unique molecular index was 9.4. The number of amplicons that could detect variants at limit of detection of 0.1% increased corresponding to the level of molecular depth (Figure S3, available online: https://cdn.amegroups.cn/static/public/tlcr-21-981-4.xlsx). The median and average of median LoD per amplicon are 0.22% and 0.20%, respectively, with the median cfTNA input at 20 ng (range, 11–36 ng) from 119 NSCLC samples (available online: https://cdn.amegroups.cn/static/public/tlcr-21-981-4.xlsx). The success rates for library construction and sequencing using the automated Genexus system were 100% (119/119) and 91% (108/119), respectively. Among the 11 samples that were failed to be sequenced, the constructed libraries were retrieved and re-sequenced using Ion Chip 540 that have equivalent reads with GX5TM chip on Ion S5 Prime system. The automated process of Genexus system from library construction, sequencing to data analyses is approximately 18 h for 4 cfTNA samples in one run.
Table 2
| Parameters | Median | Average | Range |
|---|---|---|---|
| Library input (ng) | 20 | 23.2 | 11.1–36.4 |
| Total reads | 10,467,880 | 10,369,608 | 4,765,877–15,510,927 |
| Mapped reads | 10,174,293 | 10,106,962 | 4,358,939–15,174,343 |
| Mean read length (bp) | 97 | 96 | 81–102 |
| Mean depth (×) | 35,773 | 36,094 | 13,485–55,882 |
| Median molecular coverage (×) | 2,192 | 2,141 | 590–3,311 |
| Molecular uniformity | 92% | 90% | 52–89% |
| Median reads per functional molecules | 9.4 | 9.9 | 4.6–42.3 |
| Median perc functional reads | 60% | 58% | 29–74% |
| Molecular conversion efficiency* | 32% | 32% | 10–56% |
*, calculation based on 1ng equivalent to 300 copies of genome. Evaluated using N=108 (11 samples were sequenced using Ion Chip 540 + IonS5 Prime). NGS, next generation sequencing.
Relative molecular coverage per amplicon revealed that the amplification efficiency of 14 of the 250 amplicons were underperformed with relative molecular coverage of <0.5, compared with the median molecular coverage of individual samples (Figure S4). Among the 11 genes that are related to the somatic mutations on labels and guidelines of NSCLC as well as those in TP53, relative molecular coverage of 2 amplicons, MET_SP_191.10347 and KRAS_r1_1.1.811 were below 0.5 (Figure S5). However, both of the amplicons were used for CNV calling.
Genomic alterations detected from liquid biopsy
Among the 119 NSCLC patients examined, Genexus-OPA analysis detected at least one alteration in 79 patients (66%) from plasma cfTNA (Figure 1A). Among the 130 alterations detected, 111 were SNVs or insertion/deletions (INDELs), 15 were CNV and 4 were fusion genes (Figure 1, available online: https://cdn.amegroups.cn/static/public/tlcr-21-981-5.xlsx). The most-frequently mutated genes comprised of TP53 (35%), EGFR (19%), KRAS (15%), PIK3CA (6%), BRAF (3%), CDKN2A (3%) and FGFR3 (3%) (Figure 1B). Gene amplifications were detected in EGFR, ERBB2, CDKN2A, MET, FGFR2, ERBB3 and KRAS whilst one deletion was detected on CDKN2A. Four fusion alterations were EML4-ALK (in 2 cases), SLC3A2-NRG1 (in one case) and KIF5B-RET (in one case) (Figure 1A).
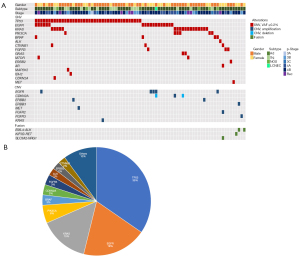
Concordance rate between tissue biopsy and liquid biopsy
The concordance rate of genomic profiles between tumor tissue biopsy and liquid biopsy was evaluated with 63 genomic alterations covered by both Genexus-OPA and SOC tissue testing. Fifty-nine percent (37 alterations—17 EGFR, 14 KRAS, 2 BRAF, 1 ERBB2, 2 ALK fusions and 1 RET fusion) of actionable genomic alterations detected in tumor tissues were also concordantly detected in plasma cfTNA (Figure 2, available online: https://cdn.amegroups.cn/static/public/tlcr-21-981-6.xlsx). A total of 26 genomic alterations were detected by SOC tissue testing only (Figure 2, available online: https://cdn.amegroups.cn/static/public/tlcr-21-981-6.xlsx). On the other hand, 5 genomic alterations, KRAS p.G12V (GNX-071), KRAS p.G12C (GNX-082), KRAS p.Q61H (GNX-129) MET p.D1028N (GNX-139) and EGFR p.L858R (GNX-145), were detected from liquid biopsy only (available online: https://cdn.amegroups.cn/static/public/tlcr-21-981-6.xlsx). Among them, EGFR p.L858R, KRAS p.G12C and MET p.D1028N were on level 1 evidence based on OncoKB Therapeutic Level of Evidence V2 (13). Level 1 evidence are FDA-recognized biomarker predictive of response to an FDA-approved drug in this indication. The sensitivity of Genexus-OPA NGS system to detect EGFR mutation with Cobas EGFR test was 49%, to detect ALK fusion with ALK-IHC was 67% and to detect 9 major gene alterations with Oncomine Dx target test was 66% (Table 3). The specificity of Genexus-OPA NGS system to detect EGFR mutation with Cobas EGFR test was 99%, to detect ALK fusion with ALK-IHC was 100% and to detect 9 major gene alterations with Oncomine Dx target test was 93% (Table 3).

Table 3
| SOC CDx assay | Genexus | |
|---|---|---|
| Positive | Negative | |
| Oncomine | ||
| Positive | 33 | 17 |
| Negative | 3 | 37 |
| Cobas EGFR | ||
| Positive | 17 | 18 |
| Negative | 1 | 75 |
| ALK-IHC | ||
| Positive | 2 | 1 |
| Negative | 0 | 103 |
Oncomine Dx examined in 89/119 cases. Sensitivity 66.0%; specificity 92.5%. Oncomine Dx target test evaluated 9 major genes in NSCLC: EGFR, ALK, ROS1, BRAF, MET, KRAS, ERBB2, NTRK and RET. GNX-139 counts two times (for EGFR and MET). Cobas EGFR examined in 111/119 cases. Sensitivity 48.57%; specificity 98.68%. ALK-IHC examined in 106/119 cases. Sensitivity 66.67%; specificity 100.0%. ctDNA, circulating tumor DNA; IHC, immunohistochemistry; NSCLC, non-small cell lung cancer.
Association between clinical parameters and ctDNA detection
The associations of cfTNA concentration and various clinical parameters with ctDNA detection were evaluated. Although we found no significant correlation between cfTNA concentration and ctDNA detection rate (Figure 3A), it is notable that patients with extrathoracic metastasis to adrenal gland, bone, and brain showed statistically-significant ctDNA detection rate (P=0.013–0.019) (Table 4). Among the routine blood test parameters, lactate dehydrogenase (LDH), C-reactive protein (CRP) and carcinoembryonic antigen (CEA) LDH, CRP and CEA were significantly higher in the patients with ctDNA detection (P=0.004–0.019) (Figure 3B).
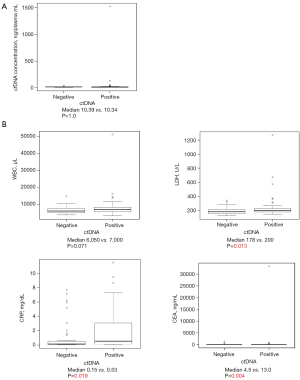
Table 4
| Parameters | ctDNA + | ctDNA − | P value |
|---|---|---|---|
| Clinical parameters | |||
| Age | 0.685 | ||
| ≥65 years | 52 | 28 | |
| <65 years | 27 | 12 | |
| Gender | 1 | ||
| Female | 29 | 14 | |
| Male | 50 | 26 | |
| Smoking | 0.513 | ||
| Never | 19 | 12 | |
| Former/current | 60 | 28 | |
| Patho | 0.829 | ||
| Ad | 58 | 28 | |
| Non-Ad | 21 | 12 | |
| T stage | 0.056 | ||
| ≤2 | 27 | 15 | |
| ≥3 | 44 | 9 | |
| N stage | 0.059 | ||
| ≤1 | 37 | 18 | |
| ≥2 | 34 | 6 | |
| M stage | 0.072 | ||
| 1a | 13 | 8 | |
| 1b/1c | 43 | 9 | |
| Stage | 0.416 | ||
| III | 15 | 7 | |
| IV | 56 | 17 | |
| Metastatic sites | |||
| Lung | 0.663 | ||
| + | 20 | 12 | |
| − | 59 | 28 | |
| Pleura | 0.424 | ||
| + | 27 | 17 | |
| − | 52 | 23 | |
| Liver | 0.058 | ||
| + | 12 | 1 | |
| − | 67 | 39 | |
| Adrenal | 0.015 | ||
| + | 11 | 0 | |
| − | 68 | 40 | |
| Bone | 0.013 | ||
| + | 32 | 7 | |
| − | 47 | 33 | |
| Brain | 0.019 | ||
| + | 14 | 1 | |
| − | 65 | 39 |
Ad, adenocarcinoma; ctDNA, circulating tumor DNA.
Discussion
Although tumor biopsy remains SOC for selecting molecular-targeted drugs for NSCLC, inadequate or low quality of tissue materials as well as complications associated with biopsy procedure are some hurdles for genomic profiling. Hence, minimally invasive liquid biopsy and rapid genomic profiling are expected to be potential options for initial genomic screening. In this study, we conducted a feasibility study to assess the detection rate of actionable alterations in cfTNA of 119 newly-diagnosed NSCLC cases using OPA with the automated Integrative Genexus System. By using this system, 66% of the patients were detected to carry at least one alteration using plasma cfTNA. According to the statement paper published from the International Association for the Study of Lung Cancer (IASLC) that reported the sensitivity of mutation detection in NSCLC related genes using liquid biopsy, Genexus-OPA revealed similar detection sensitivity ranged between 49–67% among the NSCLC related genes (14).
Among the 63 genomic alterations that were covered in both SOC tissue testing and Genexus-OPA, the Genexus-OPA could concordantly detect genomic alterations identified from SOC tissue testing in 59% cases. The concordance rate is comparable with the MSK study, which indicated 59% of concordance between MSK-IMPACT (tissue profiling) and MSK-ACCESS (liquid biopsy profiling) (15). Five genomic alterations were detected in plasma cfTNA only. The discordance could be partly explained by tumor heterogeneity. For examples, a KRAS G12C mutation was detected in plasma cfTNA of patient GNX-082 (stage IVB) but not detected with lung biopsy with Oncomine Dx. Similarly, EGFR p.L858R was detected in plasma cfTNA of GNX-145 (stage IVA) but not detected from pleural effusion sample examined with Cobas EGFR test. Importantly, both of the mutations are actionable mutations with OncoKB level 1 evidence that was critical for treatment decision. These results further highlighted liquid biopsy could be used as complementary to tumor tissue biopsies to identify actionable mutations, particularly in addressing tumor heterogeneity in metastatic or recurrence tumors. In addition, the discordance might also cause by the possibility of detecting clonal hematopoiesis related mutations from plasma that were not detected in tumor profiling (16).
Additionally, Genexus-OPA system has advantage for its quick TAT. For example, in the case of GNX-074, StageIVB NSCLC patient, genomic profiling of liquid biopsy was performed by Genexus-OPA. On the same day, a bone biopsy sample was prepared for Oncomine Dx Target test. Genexus-OPA system found a KRAS G12C mutation within 4 days. Eight days later, the result of Oncomine Dx Target (SOC test) identified the same mutation from the bone biopsy sample. Several factors that could affect the TAT on comprehensive genomic profiling that include (I) laborious procedure of NGS, (II) waiting period to collect optimal number of samples to be pooled for sequencing, (III) sample logistic in which samples would have to be delivered to overseas distributor if local service is not available. All the aforementioned factors were addressed by the Integrative Genexus System emphasizing the flexibility, quick TAT and the potential feasibility of in-house genomic profiling. Genexus-OPA required only 18 h to complete library construction, templating, sequencing and data analyses. The shorter TAT approach provided by Genexus-OPA suggested the possibility of performing cfDNA genomic profiling in advanced to detect actionable mutations while waiting for the results of tumor biopsy. However, one of the shortcomings of Genexus-OPA system is the relatively low detection of EGFR mutations (17/35) that required further improvement of primer designs in the panel.
In this study, we also investigated various clinicopathological and biological parameters that might affect ctDNA detection rates with liquid biopsy. As for biological parameter, Li et al. reported the association of cfDNA concentration and increased ctDNA detection, however we found no significant association (17). Among the clinical parameters, advanced tumor stages and metastatic status are known to be associated with ctDNA detection (18,19). In this study, extra-thoracic metastasis (adrenal gland, bone and brain metastasis) was one of significant parameters to be related to ctDNA detection rates as reported in previous reports (18,19). Additionally, LDH, CRP and CEA levels were found to be significantly associated with the ctDNA detection rate with liquid biopsy. LDH and CEA levels increase corresponding to the tumor burden that is known to correlated with ctDNA levels (20,21). CRP is known to represent inflammation in the body. Further study is necessary to identify the factors for predicting ctDNA detection under inflammation states in either primary or metastatic sites.
In conclusion, a feasibility study was conducted to evaluate the sensitivity and specificity of genomic alterations in liquid biopsy using a fully automated next-generation sequencer (Ion Torrent™ Genexus™ Integrated Sequencer) and OPA in comparison of tissue biopsies.
Acknowledgments
We thank the patients and their families for their participation in the study. We thank the staff of the Department of Thoracic Medical Oncology, Cancer Institute Hospital, Japanese Foundation for Cancer Research, for supporting this study. We would like to convey gratitude to ThermoFisher Scientific technical support team for their continuous support in this study.
Funding: This work was supported by the Council for Science, Technology and Innovation (CSTI), cross-ministerial Strategic Innovation Promotion Program (SIP), “Innovative AI Hospital System” Funding Agency, National Institute of Biomedical Innovation, Health and Nutrition (NIBIOHN) and JSPS KAKENHI (No. 20K07709).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-21-981/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-21-981/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-21-981/coif). SKL serves as a scientific advisor and received honorarium from Cancer Precision Medicine Inc. RA receives personal fees for lectures from AstraZeneca, Chugai Pharmaceutical. KU receives personal fees for lectures from AstraZeneca, Brystol Myers Squibb, Chugai Pharmaceutical, Daiichi Sankyo, Eli Lilly, Novartis, Ono Pharmaceutical, and Takeda Pharmaceutical Company Limited, and for manuscript writing from Chugai-Igakusha, Nankodo Co., Ltd., Nanzando Co., Ltd., Yodosha Co., Ltd. KU’s spouse is an employee of Daiichi Sankyo. RH and YMC are Employees of Cancer Precision Medicine, Inc. NY is a medical advisor of Chugai Pharmaceutical and receives lecture’s fee from Ono Pharmaceutical, Bristol Myers Squibb, Pfizer, Chugai Pharmaceutical, Eli Lilly, Taiho Pharmaceutical, AstraZeneca, MSD, and Takeda Pharmaceutical Company Limited. YN serves as a scientific advisor of Oncotherapy Science Inc., and possesses stock from Oncotherapy Science Inc. MN receives grants and personal fees from Ono Pharmaceutical, Bristol Myers Squibb, Pfizer, Chugai Pharmaceutical, Eli Lilly, Taiho Pharmaceutical, AstraZeneca, MSD, Novartis, Daiichi Sankyo, and Takeda Pharmaceutical Company Limited, personal fees from Boehringer-Ingelheim, Merck Biopharma, Teijin Pharma Limited, and AbbVie. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study design is approved by the ethical committee in Japanese Foundation for Cancer Research (No. IRB-2020-1055) and all patients provided written informed consent to this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- Lin JJ, Cardarella S, Lydon CA, et al. Five-Year Survival in EGFR-Mutant Metastatic Lung Adenocarcinoma Treated with EGFR-TKIs. J Thorac Oncol 2016;11:556-65. [Crossref] [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Ramalingam SS, Vansteenkiste J, Planchard D, et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N Engl J Med 2020;382:41-50. [Crossref] [PubMed]
- Mok T, Camidge DR, Gadgeel SM, et al. Updated overall survival and final progression-free survival data for patients with treatment-naive advanced ALK-positive non-small-cell lung cancer in the ALEX study. Ann Oncol 2020;31:1056-64. [Crossref] [PubMed]
- Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311:1998-2006. [Crossref] [PubMed]
- Meric-Bernstam F, Brusco L, Shaw K, et al. Feasibility of Large-Scale Genomic Testing to Facilitate Enrollment Onto Genomically Matched Clinical Trials. J Clin Oncol 2015;33:2753-62. [Crossref] [PubMed]
- Sundaresan TK, Sequist LV, Heymach JV, et al. Detection of T790M, the Acquired Resistance EGFR Mutation, by Tumor Biopsy versus Noninvasive Blood-Based Analyses. Clin Cancer Res 2016;22:1103-10. [Crossref] [PubMed]
- Villaflor V, Won B, Nagy R, et al. Biopsy-free circulating tumor DNA assay identifies actionable mutations in lung cancer. Oncotarget 2016;7:66880-91. [Crossref] [PubMed]
- Heitzer E, Haque IS, Roberts CES, et al. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet 2019;20:71-88. [Crossref] [PubMed]
- Hayashi H, Tanishima S, Fujii K, et al. Clinical impact of a cancer genomic profiling test using an in-house comprehensive targeted sequencing system. Cancer Sci 2020;111:3926-37. [Crossref] [PubMed]
- Kou T, Kanai M, Yamamoto Y, et al. Clinical sequencing using a next-generation sequencing-based multiplex gene assay in patients with advanced solid tumors. Cancer Sci 2017;108:1440-6. [Crossref] [PubMed]
- Chakravarty D, Gao J, Phillips SM, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017; [Crossref] [PubMed]
- Rolfo C, Mack PC, Scagliotti GV, et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J Thorac Oncol 2018;13:1248-68. [Crossref] [PubMed]
- Rose Brannon A, Jayakumaran G, Diosdado M, et al. Enhanced specificity of clinical high-sensitivity tumor mutation profiling in cell-free DNA via paired normal sequencing using MSK-ACCESS. Nat Commun 2021;12:3770. [Crossref] [PubMed]
- Chan HT, Nagayama S, Chin YM, et al. Clinical significance of clonal hematopoiesis in the interpretation of blood liquid biopsy. Mol Oncol 2020;14:1719-30. [Crossref] [PubMed]
- Li BT, Janku F, Jung B, et al. Ultra-deep next-generation sequencing of plasma cell-free DNA in patients with advanced lung cancers: results from the Actionable Genome Consortium. Ann Oncol 2019;30:597-603. [Crossref] [PubMed]
- Aggarwal C, Thompson JC, Black TA, et al. Clinical Implications of Plasma-Based Genotyping With the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA Oncol 2019;5:173-80. [Crossref] [PubMed]
- Karlovich C, Goldman JW, Sun JM, et al. Assessment of EGFR Mutation Status in Matched Plasma and Tumor Tissue of NSCLC Patients from a Phase I Study of Rociletinib (CO-1686). Clin Cancer Res 2016;22:2386-95. [Crossref] [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [Crossref] [PubMed]
- Jovelet C, Ileana E, Le Deley MC, et al. Circulating Cell-Free Tumor DNA Analysis of 50 Genes by Next-Generation Sequencing in the Prospective MOSCATO Trial. Clin Cancer Res 2016;22:2960-8. [Crossref] [PubMed]


