
Triptolide promotes degradation of the unfolded gain-of-function Tp53R175H/Y220C mutant protein by initiating heat shock protein 70 transcription in non-small cell lung cancer
Introduction
Lung cancer is most common cause of cancer-related death, accounting for 18.4% of cancer deaths worldwide (1). Among the two histological subtypes of lung cancer, non-small cell lung cancer (NSCLC) accounts for approximately 85% of the incidence rate worldwide (2). According to data obtained from The Cancer Genome Atlas (TCGA) database, 41.8% of cancer patients have altered tumor protein P53 (TP53) (3), and the mutation rate of TP53 in NSCLC is 50%, which suggests that mutant P53 (mutp53) plays an important role in NSCLC progression.
TP53, which encodes the tumor-inducing protein P53, is one of the most tumor suppressor genes (4,5). Functionally, p53 binds directly to chromatin and plays an important role in regulating the cell cycle, apoptosis, autophagy and DNA repair in response to damaging factors (6). However, mutations of TP53 usually alter the structure of the DNA-binding domain (preventing protein folding) or affect the residues directly in contact with DNA by causing folding error, and missense mutations in the DNA-binding domain often lead to changes in the three-dimensional structure of p53, leading to a dominant-negative phenotype with a diminished ability to transactivate target genes (7). Mutated p53 proteins alter cellular programs that accelerate tumor progression and metastasis by physically recruiting other transcription factors, reprogramming cell behavior, promoting cancer cell survival and proliferation, invasion and migration, stem cell renewal, chemoresistance, and matrix remodeling, exhibiting gain-of-function (GOF) features (8-12). A growing body of literature suggests that TP53 mutations in lung cancer are associated with increased resistance to cancer therapy and poorer survival prognosis (6), thus, mutp53 is a promising therapeutic target. Many natural or naturally derived antitumor drugs [curcumin (13), artemisinin (14), capsaicin (15)] have begun to play increasingly important roles in clinical cancer treatment (16).
Triptolide is an epoxy diterpene lactone that has been extracted from Tripterygium wilfordii, a vine widely distributed in southern China that has been used for centuries to treat inflammation and autoimmune diseases (17). Triptolide has a strong killing effect on breast (18), prostate (19), pancreatic (20), gastric (21), and colon (22,23) cancer cells but is limited in the clinic due to toxicity. Study has indicated that triptolide inhibits the migration and proliferation of fibroblasts from ileocolonic anastomosis of patients with Crohn’s disease by regulating the microRNA (miRNA) miR-16-1/heat shock protein 70 (HSP70) pathway (24). However, the mechanism of triptolide action in the treatment of metastatic NSCLC with mutp53 expression is still unclear.
HSPs are sensors and regulators of conformational protein equilibrium that maintain the activity, stability, and integrity of the cell proteome and intracellular transport for overall dynamic cellular balance (19,25-29). Under normal circumstances, HSP90 binds to P53 to inhibit murine double minute 2 homolog (MDM2) promotion of P53 protein ubiquitination and degradation. However, study has shown that the unfolded mutp53 protein or inhibited HSP90 activity causes the unstable unfolded mutp53 to bind preferentially to HSP70, which can promote mutp53 degradation by the proteasome (30).
In this study, we discovered a previously unknown mechanism by which triptolide leads to rapid and selective depletion of unfolded mutp53 proteins through the ubiquitin-proteasome system. Using short hairpin RNA (shRNA)-mediated gene knockdown, we found that HSP70 plays a significant role in the triptolide regulation of TP53R175H/Y220C mutant expression in NCI-H1299 cells. In summary, our study showed that triptolide can specifically degrade unfolded mutp53 proteins and holds the potential to become a specific therapeutic drug targeting unfolded mutp53 in NSCLC. We present the following article in accordance with the ARRIVE reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-312/rc).
Methods
Cell lines, cell culture, and reagents
Human lung epithelial NCI-H1299 (TP53-null) and HEK-293T cells were purchased from Shanghai Fuheng Biological (Shanghai, China). All cell lines were authenticated by short tandem repeat (STR) profiling identification. New cell batches were thawed every 3–4 months for use in these experiments. NCI-H1299 cells were maintained in RPMI-1640 (Corning Inc., Corning, NY, USA) medium, and HEK-293T cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM; Corning). Both media contained 10% fetal bovine serum (FBS; Corning) and 1% penicillin/streptomycin (Solarbio, Beijing, China), and all cells were maintained at 37 ℃ in a humidified chamber with 5% CO2 and passaged twice weekly. The agent triptolide was purchased from MCE (New Jersey, USA) and used at a concentration of 30 nM (the dose selection of triptolide refers to the previous experimental research in this laboratory (31). After pre-experiment, the drug concentration and drug action time of 30 nmol, which has no killing effect on cells and has the most obvious effect, are selected). MG132 was used at a concentration of 20 µM, and nutlin-3A was used at a concentration of 20 µM. Matrigel was purchased from BD (356234, Franklin Lakes, NJ, USA).
Plasmids
To overexpress TP53, a plasmid expressing wild-type P53 (wtp53) was purchased from WZ Biosciences Inc (Shandong, China). The polymerase chain reaction (PCR) product was cloned into a pENTER (WZ Biosciences Inc., Shanghai, China) vector backbone. The cytomegalovirus (CMV)-TP53Y220C/R273H/H179L/R248Q/H193R plasmid was generated with a Quick Mutation™ Plus site-directed mutagenesis kit (D0208S, Beyotime Biotechnology, Shanghai, China) using CMV-wtp53 as a template. Products were validated by Sanger sequencing to authenticate the mutations and ensure the quality of the constructs. Plasmids were transfected into NCI-H1299 cells using Lipofectamine 3000TM (Invitrogen, Waltham, MA, USA) according to the manufacturer’s instructions. The sequences of the mutant plasmids are listed in Table S1.
Lentivirus construction and cell transfection
The TP53R175H plasmid was purchased from the Miaoling Plasmid Platform (miaolingbio.com). To knock down HSP70 expression, 3 fragments targeting the sequences shHSPA1A-1 (GGA TCC AGT GTT CCG TTT), shHSPA1A-2 (GTC AGT TCT CAA TTT CCT GTG), and shHSPA1A-3 (GCC ATC TTA CGA CTA TTT CTT) were synthesized by TSINGKE BioTech (Qingdao, China), and shHSP70 was constructed through annealing, restriction digestion, and enzyme linking and subcloned into a pGIPZ vector. After sequencing verification, the lentiviruses were cotransfected in a shHSP70/TP53R175H plasmid with the packaging plasmids pSPAX2 and pMD2G. Cell transfection was performed with Hitrans GP (GeneChem, Shanghai, China) in accordance with the manufacturer’s protocols, and clones were selected with puromycin (1–2 µg/mL) 10–14 days after transfection.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cell lines using trizol (Invitrogen). For qRT-PCR, 1 µg of RNA was directly reverse transcribed using an lnRcute lncRNA First-Strand complementary DNA (cDNA) synthesis kit [with genome DNase (gDNase); KR202, TIANGEN BIOTECH Co., Ltd., Beijing, China] containing oligo(dT) primers, and RNA expression was evaluated by qRT-PCR with an lnRcute lncRNA qPCR kit (SYBR Green, FP402-02, TIANGEN BIOTECH) and a QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA). Human primers were used in this study. Beta-actin (ACTB) was used as the internal control, and the fold change in target gene expression was calculated by the delta delta Ct method. All data are mean ± standard deviation (SD). Each experiment was repeated at least 3 times (32,33). The primers are listed in Table S2.
EdU assay
For the 5-ethynyl-2'-deoxyuridine (EdU) staining (C0078S, Beyotime Biotechnology) analysis, 4×103 cells were seeded on a 96-well plate and incubated overnight. Then, an EdU working solution was added, and the cells were fixed with 4% paraformaldehyde for 15 minutes at room temperature. According to the dye manufacturer’s protocol, after washing and blocking the cells with 3% bovine serum albumin (BSA) and 0.3% Triton X-100, 1X Hoechst was used to stain nuclei. Images were captured using a fluorescence microscope (DMi8 Inverted fluorescence microscope, Leica Microsystems, Wetzlar, Germany). The percentage of EdU-positive cells was calculated as follows: (EdU-stained cells/Hoechst-stained cells) × 100%. All data are mean ± SD. Each experiment was repeated at least 3 times (32,33).
Wound healing assay
A cell suspension was prepared and diluted to the appropriate concentration {[3–7]×105 cells/mL}, and then 110 µL of the cell suspension was added to each well containing a culture insert (80460, ibidi, Martin Reid, Germany) and incubated overnight at 37 ℃ in a humidified chamber with 5% CO2. When the cell growth density reached approximately 95%, sterile tweezers were used to grab 1 side of the insert (which had been scratched to create a wound). Subsequently, it was removed, and was placed quickly in the serum-free medium. The width of the wound was photographed using microscopy (100X, CKX53, Olympus, Shanghai, China), and the changes in width were calculated using ImageJ software (National Institutes of Health, Bethesda, MD, USA), with the results normalized to those of the control group. All data are mean ± SD. Each experiment was repeated at least 3 times (32,33).
Cell migration and invasion assays
In summary, 4×104 cells were suspended in 200 µL of serum-free medium and added to the upper chamber of each insert, which was coated with Matrigel, and 600 µL of medium supplemented with 10% FBS was added to the lower chamber of each insert. After incubation for 48 hours at 37 ℃ with 5% CO2, the cells were fixed with 4% paraformaldehyde and methanol and stained with 1% crystal violet. The cells that passed through the insert membrane were counted under an inverted microscope (CKX53, Olympus). The migration experiment was performed in the same way as the invasion experiment except for the incubation time which was 24 hours and for the inserts which were not covered in Matrigel. All data are mean ± SD. Each experiment was repeated at least 3 times (32,33).
Luciferase reporter assay
NCI-H1299 cells were seeded in 6-well plates and cotransfected with a mixture of 2.5 µg of p-Renilla luciferase-thymidine kinase (pRL-TK), PLG3-basic luciferase and P53 mutation plasmids. After 48 hours of incubation, the cells were treated with triptolide (30 nM for 0, 12, 24, 36, or 48 hours), and then firefly and Renilla luciferase activities were quantified with a dual-luciferase reporter assay kit (Promega, Madison, WI, USA) and a Cytation 5 Cell Imaging Multi-Mode Reader (BioTek Instruments, Winooski, VT, USA). The results were normalized using the internal TK signal. All data are mean ± SD. Each experiment was repeated at least 3 times (32,33).
Western blot analysis
Cells were lysed in RIPA lysis buffer (Beyotime Biotechnology), and 20 µg of protein was separated via 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA, USA). After blocking with 5% milk and incubation with primary antibodies against TP53 (1:1,000, Santa Cruz Biotechnology, Dallas, TX, USA), HSP70 (1:1,000, Proteintech, Wuhan, China), glyceraldehyde 3-phosphate dehydrogenase (GAPDH, 1:5,000, Cell Signaling Technology, Beverly, MA, USA), and ACTB (1:5,000, ABclonal Technology, Wuhan, China) at 4 ℃ overnight, the membranes were incubated with secondary antibodies (1:5,000) at room temperature for 1–2 hours. Then, the bands were examined with an Immobilon™ western chemiluminescent horseradish peroxidase (HRP) substrate (Millipore) and an Anon 5200 system (Tanon Science & Technology Co. Ltd., Shanghai, China). GAPDH or ACTB was used as the loading control.
Immunofluorescence analysis
NCI-H1299R175H/Y220C (1×105) cells were seeded into confocal dishes for 24 hours and then divided into control group and triptolide (30 nM, 24 hours) treatment group. The treated cells were fixed with 4% paraformaldehyde for 15 minutes and then permeabilized with 0.5% Triton-100 at room temperature for 20 minutes. After blocking with goat serum for 30 minutes at room temperature, 2 diluted primary antibodies (anti-TP53, mouse, 1:200 and anti-HSP70, rabbit, 1:200) were incubated overnight at 4 ℃. Fluorescent secondary antibody and 4',6-diamidino-2-phenylindole (DAPI) were added on the second day for confocal imaging.
TCGA database
Sample information was acquired from TCGA. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and Gene Set Enrichment Analysis (GSEA) were performed with the Bioconductor “clusterProfiler” package (34) and SangerBox analysis tools (http://sangerbox.com/Tool). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Lung metastasis models
Animal experiments were performed under project license (No. KYLL-2018(KJ)A-0046) granted by the Ethics Committee for Animal Experimentation of The Second Hospital of Shandong University, in compliance with Institutional Animal Care and Use Committee guidelines. Sixteen male BALB/c nude 4-week-old mice were purchased from Vital River Research Animal Services (Beijing, China) and raised in a specific pathogen-free (SPF) environment with adequate food and water (light-dark cycle: 12–12 hours, 22–25 ℃), and the litter was replaced every 3 days. For the metastatic cancer model, the TP53R175H cells [2×106 cells in 100 µL of phosphate buffered saline (PBS)] were injected into the tail vein of the nude mice (5–6 weeks old, male, 18–20 g). One week later, the mice were randomly allocated into 2 groups (8 mice per group) and then subjected to treatment. The reagents were intraperitoneally injected as follows: (I) 100 µL solvent [PBS/dimethyl sulfide (DMSO) =19:1]; and (II) triptolide (0.6 mg/kg). The body weight of the mice was measured every 3 days to determine whether triptolide had toxic effects. If the weight loss exceeded 20% or the mice become unable to eat, euthanasia was performed. Ten weeks after the first injection, the mice were sacrificed by continuous inhalation with 30% CO2 for 5 minutes, and the lung tissues were removed and divide into 2 parts. One part was fixed in 10% paraformaldehyde buffer for further analysis, and the other part was stored at −80 ℃ for later use.
Statistical analysis
Data were analyzed using a Student’s t-test with GraphPad 7.0 (GraphPad Software Inc., San Diego, CA, USA). Each experiment was repeated at least 3 times. All results are presented as the means ± SD. A P value less than 0.05 was considered statistically significant.
Results
TP53R175H/Y220C proteins significantly promoted NCI-H1299 cell proliferation, migration, and invasion
Many recent studies have shown that mutp53 is associated with human cancer cell invasion and migration (26-29). We first investigated whether unfolded mutp53 acted as a tumor-promoting gene in human NSCLC cells. Specifically, after overexpressing TP53R175H/Y220C (Figure 1A and Figure S1A), NCI-H1299 cells exhibited enhanced migratory capacity, as demonstrated by the wound healing assay (PWT =0.0209, PY220C =0.0444, PR175H =0.0127) and EdU (PWT =0.0352, PY220C =0.0078, PR175H =0.0073) experiment results (Figure 1B,1C and Figure S1B,S1C). Moreover, the transwell assay demonstrated that TP53R175H/Y220C significantly promoted NCI-H1299 cell migration (PWT =0.0006, PY220C =0.0007, PR175H =0.0021) and invasion (PWT =0.0002, PY220C =0.0036, PR175H =0.0034) in comparison to the control and TP53WT cells (Figure 1D and Figure S1D). Notably, the results indicated that TP53R175H/Y220C promoted NSCLC cell proliferation and metastasis in vitro. To assess the in vivo effect of unfolded mutp53 on lung metastasis, a lung metastasis mouse model was generated. At the end of the animal study, 14 mice were included and completed (n=7 per group). Two animals were excluded, including 1 mouse with weight loss that exceeded 20% and another that became unable to eat. The nude mice were injected with mutp53R175H overexpressing cells (which we had constructed with lentivirus carrying the TP53R175H mutant; Figure S1E and treated with triptolide, which has no toxic effects on mice (Figure S1F), and fewer metastatic foci were generated in the mice treated with triptolide than in the untreated mice (Figure 1E). In addition, the transwell assay demonstrated that triptolide had the same effect in vitro as it did in vivo (Figure 1F).
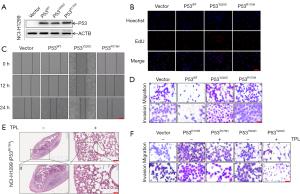
Triptolide specifically degraded unfolded GOF mutp53 proteins
We found that the natural compound triptolide could inhibit the metastasis of mutp53-expressing NSCLC cells. To investigate the effect of triptolide on unfolded mutp53 proteins, we constructed mutp53-expressing cells (Figure S1G) and discovered that triptolide caused a rapid and sustained decrease in the levels of unfolded GOF mutp53R175H/Y220C/R179L proteins, which are usually stably expressed in NSCLC (Figure 2A and Figure S1H). Interestingly, in contrast to the considerable effect on mutp53R175H/Y220C/R179L expression, triptolide exerted a negligible influence on the level of wtp53 protein in NSCLC cells (Figure 2B and Figure S1H). Furthermore, NSCLC treatment exhibited minimal effects on the other frequently observed folded GOF mutp53 protein (R248Q, H193R; Figure 2C and Figure S1H) but did cause folded mutp53R273H protein degradation in the NCI-H1299 cell line (Figure 2D and Figure S1H). Therefore, we believe that triptolide specifically degraded unfolded mutp53 proteins and selectively degraded folded mutp53 proteins. To explore the mechanism by which triptolide leads to the rapid degradation of unfolded mutp53 proteins, we performed confocal microscopy and found that triptolide caused mutp53R175H/Y220C protein translocation from the nucleus to the cytoplasm, which induced the degradation of the mutp53 protein (Figure 2E).
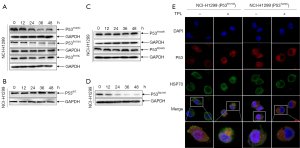
Triptolide induced mutp53R175H/Y220C through proteasome degradation mediated by the MDM2 ubiquitin ligase
To determine whether triptolide regulates wtp53 and mutp53 expression at the transcriptional level or posttranscriptional level, we treated NCI-H1299 cells with triptolide and the proteasome inhibitor cycloheximide (CHX). The results showed that triptolide caused a reduction in the half-life of stable mutp53R175H/Y220C proteins (Figure 3A). Interestingly, in contrast to the effect on mutp53R175H/Y220C proteins, triptolide exerted a negligible influence on the level of wtp53 proteins in NSCLC cells (Figure 3B), indicating that the effect of triptolide on mutp53WT/R175H/Y220C was posttranslational. Because triptolide caused increased mutp53R175H/Y220C degradation, we sought to determine whether the effect was mediated by the proteasome. The proteasome inhibitor MG132 rescued mutp53R175H/Y220C proteins from triptolide-induced degradation (Figure 3C and Figure S2A, PR175H =0.0428, PY220C =0.0009), suggesting that the protein is targeted by the proteasome for degradation. To confirm this result, we ectopically expressed His-tagged ubiquitin (His-Ub) in NCI-H1299 cells, and treatment with triptolide caused ubiquitination of mutp53R175H/Y220C (Figure 3D). Because MDM2 played a vital role in the degradation of the mutp53 protein, nutlin3 was used to detect the mechanism by which the mutp53 protein was degraded and to determine whether this mechanism was mediated by the MDM2 E3 ubiquitin ligase. The results indicated that triptolide-induced mutp53R175H/Y220C protein degradation (PR175H =0.0121, PY220C =0.0339) was mediated by the MDM2 ubiquitin ligase (Figure 3E and Figure S2B).

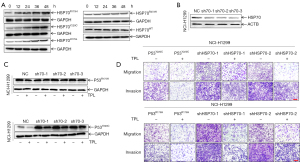
Triptolide regulated the HSP70-mediated degradation of unfolded GOF mutp53 proteins
The stability of mutp53 is inseparable from the interaction of its molecular chaperones, including HSP70 and HSP90 (35). Previous experiments have shown that HSP90 can bind to folded mutp53 proteins, inhibiting MDM2 promotion of TP53 ubiquitination and resulting in increased stability of these mutp53 proteins (36,37), leading to their cellular accumulation. However, HSP70 can specifically interact with unfolded mutp53R175H/Y220C proteins and target them for degradation (30). To further study the mechanism by which HSP70 influences triptolide regulation of unfolded mutp53 protein degradation, we treated mutp53 with triptolide in a dose-dependent manner. We found that HSP70 expression was significantly increased with triptolide treatment of unfolded mutp53-expressing cells but not with that of wtp53-expressing cells (Figure 4A). To determine whether HSP70 is specifically required for triptolide-induced unfolded mutp53 protein degradation, we used 3 shRNAs to target HSP70 and determined the transfection efficiency by western blotting (WB; Figure 4B). We noticed that shHSP70 partially reversed triptolide-induced mutp53R175H/Y220C degradation [PR175H(sh70-1/2/3) =0.0232/0.0295/0.0101, PY220C(sh70-1/2/3) =0.0232/0.1412/0.0410] (Figure 4C and Figure S2C). In addition, transwell assays were performed to identify the roles of HSP70 in mutp53R175H/Y220C expressing NSCLC cell metastasis [PY220C(sh70-1/2) =0.002/0.0011, PR175H(sh70-1/2) =0.0442/0.0224] and migration [PY220C(sh70-1/2) =0.0269/0.0097, PR175H(sh70-1/2) =0.002/0.0011] (Figure 4D and Figure S2D,S2E). These data indicated that triptolide regulated the HSP70-induced degradation of unfolded GOF mutp53 protein.
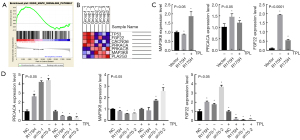
NCI-H1299R175H influenced mitogen-activated protein kinase (MAPK) pathway activation
Previous experiments have shown that mutp53R175H enhances the proliferation, migration, and invasion of NCI-H1299 cells in vitro and in vivo. To further explore the downstream genes affected by mutp53R175H, we used the University of California, Santa Cruz (UCSC) Xena (https://xena.ucsc.edu/) website to download lung adenocarcinoma data from TCGA. Online SangerBox tools were used to identify mutp53R175H and TP53STOP samples (which indicated that TP53 does not undergo transcription) from more than 500 lung adenocarcinoma samples. More than 20,000 differentially expressed genes were identified through a volcano map analysis, and these differentially expressed genes were further assessed using KEGG pathways and GSEA. We found that these differentially expressed genes may be involved in regulating the MAPK pathway (Figure 5A,5B); therefore, we selected some key differentially expressed genes in the MAPK signaling pathway for verification and assessed the treatment effects of triptolide on their expression. The results showed that the treatment effects on the protein kinase CAMP-activated catalytic subunit alpha (PRKACA) (P=0.0159), fibroblast growth factor 22 (FGF22) (P<0.0001), and mitogen-activated protein kinase kinase kinase 8 (MAP3K8) (P=0.0223) genes were consistent with the enrichment results and that triptolide reversed the cellular effects induced by mutp53R175H (Figure 5C,5D) (PPRKACA =0.0479, PFGF22 <0.0001, PMAP3K8 =0.002). These findings suggested that mutp53R175H might play a cancer-promoting role by regulating the MAPK signaling pathway. Furthermore, shHSP70 reversed the effects induced by p53R175H expression (Figure 5D).
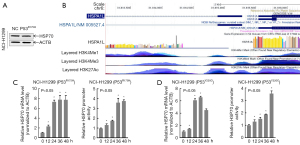
Triptolide increased HSP70 promoter activity in a time-dependent manner
We verified that there was no direct upstream or downstream relationship between HSP70 and TP53 (Figure 6A). To explore the mechanism by which triptolide increases HSP70 expression in NSCLC, we first analyzed the UCSC Genome Bioinformatics Site (http://genome.ucsc.edu/). We found that enriched levels of differentially expressed HSP70 overlapped with H3K4me1/3 (histone H3 lysine 4 monomethyl and histone H3 lysine 4 trimethyl) and H3K27Ac (Histone H3 acetylated lysine 27) peaks at the HSP70 promoter (Figure 6B). Next, the luciferase reporter gene driven by HSP70 promoter was transfected into H1299R175H/Y220C cells to evaluate the up-regulation of HSP70 mRNA transcription by triptolide. As shown in Figure 6C,6D, triptolide treatment increased HSP70 promoter activity (PR175H =0.0012, PY220C =0,0034) in a time-dependent manner. Taken together, these data confirmed that triptolide caused mutp53R175H/Y220C protein degradation by increasing HSP70 expression, which was mediated by increased HSP70 promoter activity.
Discussion
In this study, we showed that the expression of GOF mutp53R175H/Y220C proteins can enhance the invasion, migration, and proliferation abilities of NSCLC cells. At the same time, we found that triptolide can increase the transcriptional activity of HSP70, thereby increasing its protein expression, specifically degrading unfolded GOF mutp53 proteins, selectively degrading folded GOF mutp53 proteins, and ultimately inhibiting the progression of NSCLC. Therefore, we believe that a strategy to achieve targeted depletion of mutp53 proteins holds high therapeutic potential.
Previous studies have shown that GOF mutp53 proteins in a mouse model of NSCLC are associated with a high frequency of cell metastasis and invasion (20,21), TP53 is the most commonly mutated gene in human cancers. It encodes a transcription factor with a tumor-suppressing function, which it realizes by binding directly to the DNA-binding domain of a target gene, thereby limiting the accumulation of reactive oxygen species, promoting autophagy, and preventing stem cell self-renewal and other cellular processes. Moreover, a high frequency of TP53 mutations has been found in certain tumor types, such as head and neck cancer (67%), esophageal gland cancer (65%) and lung squamous cell carcinoma (84%) (38). In lung cancer-related studies, it was found that TP53 mutation can be used as a prognostic factor in patients with local-stage NSCLC (stage I–IIIA) (39), and patients with TP53 and EGFR co-mutation have relatively shorter PFS (progression free survival) compared with patients without TP53 gene mutation and resistance to EGFR tyrosine kinase inhibitors is also relatively high (40,41). Especially in the high TMB (tumor mutation burden) population, regardless of PD-L1 expression, the PFS of immunotherapy combined with immunotherapy was superior to chemotherapy, that is, the higher the tumor TMB, the more neoantigens produced by the tumor, the stronger the T cell and antitumor responses, the more PD-L1 treatment works better (42). Besides, French researchers reported that TP53 mutation status was associated with overall survival (OS) benefit in advanced NSCLC patients treated with immune checkpoint inhibitor (ICI), so after PD-L1, TMB gradually emerging as potential markers to predict the efficacy of immune checkpoint therapy (43), specific TP53 mutants also have potential as biomarkers for ICIs in NSCLC in future studies (44). Besides, in silico simulation data can provide insights into structural changes of NSCLC cells which enable invasion, migration, and proliferation (45). In lucking of structural data in silico modeling may give better view of altering the hydrogen-bonding network in the DNA-binding surface due to mutp53, compromising the direct contacts to DNA backbone phosphates due to the shorter lateral chains. Studies have shown that GOF mutp53 proteins participate in the occurrence and development of many malignant tumors by enhancing cell proliferation and tumor formation, invasiveness, and angiogenesis, promoting the tumor cell epithelial-mesenchymal transition, and endowing cancer cells with stem cell characteristics (46-50). Recently, statins have been shown to degrade mutp53 proteins through the mevalonate pathway, and zinc-pentahydroxyphenone compounds have been shown to reactivate wtp53 activity. However, most of these “reactivators” are still in the preclinical testing stage (51-53). In addition, the specific mechanism of triptolide-induced GOF mutp53 degradation is not fully understood and needs to be further explored.
Herein, we reported the use of the NCI-H1299 cell line (TP53-null) to construct R175H lentivirus-expression cells and transiently transfected Y220C plasmid cells to verify how a natural compound called triptolide influences mutp53 protein function in NSCLC. We showed that triptolide exerts different effects on the levels of wtp53 and unfolded mutp53R175H/Y220C protein expression in NSCLC cells (Figure 2A,2B). Specifically, triptolide treatment results in rapid degradation of unfolded mutp53 proteins, whereas it has a negligible effect on the half-life of wtp53 proteins. Although folded mutp53R248Q/H193R proteins are not affected by triptolide treatment, the level of folded mutp53R273H proteins is decreased after triptolide treatment (Figure 2C,2D), and the degree of this effect is consistent with that observed for unfolded mutp53 proteins, providing evidence for the specific degradation of unfolded mutp53 proteins and the selective degradation of folded mutp53 proteins by triptolide. Our results indicate that triptolide regulates unfolded mutp53 protein degradation through distinct mechanisms involving nuclear export, ubiquitination, and targeted degradation by the proteasome (Figure 2E). In our experiment, nutlin-3A rescued the triptolide-induced degradation of mutp53R175H/Y220C proteins (Figure 3C,3E). We also confirmed that mutp53 protein degradation is mediated by the ubiquitin ligase MDM2. We observed that mutp53 directly interacts with HSP70 and is important for mutp53 protein stabilization. Our results suggest that triptolide upregulates HSP70 protein expression, inducing unfolded mutp53 protein degradations in a ubiquitin-proteasome pathway-dependent manner (Figure 3C,3E), although we cannot rule out the possible effects of other underlying mechanisms.
When we introduced a shRNA targeting HSP70, we observed increased unfolded mutp53 protein expression in NSCLC cells, indicating that HSP70 plays an important role in triptolide-induced degradation of unfolded mutp53 proteins (Figure 4C). Although we identified HSP70 as an intermediate carrier for triptolide in the regulation of unfolded mutp53 protein expression, other reports have suggested that HSP40 can modulate HSP70 ATPase activity, causing the substrate polypeptide bound by fold-inducing HSP70/HSP40 to promote local mutp53 DNA-binding domain unfolding, which causes TP53 dissociation through its DNA-binding loop and leads to a potential conformational change in wtp53 (54). Similarly, in 2019, a research team at the Kalodimos Lab of St. Jude Children’s Research Hospital (55) found that the binding of HSP70 and HSP40 replaced the unfolded substrate and that HSP40 activity was changed after it was bound to HSP70, thereby further regulating the binding and release of the substrate. Therefore, we speculate that the degradation of mutp53R175H/Y220C may be closely related to HSP40 (HSP40 can regulate several intracellular proteins and signaling pathways) (56). In our study, we also found that the triptolide-mediated degradation of mutp53R175H/Y220C proteins is similar to the natural degradation of wtp53 proteins and depends on MDM2. This suggests that triptolide may induce a HSP70-mediated conformational change in mutp53R175H/Y220C mutants, rendering them similar to the wtp53 protein and thus leading to their ubiquitination and degradation. These possible roles played by HSP40 are the subjects of future research.
In summary, our study revealed that triptolide suppresses NSCLC cell invasion and migration by increasing HSP70 transcriptional activity and promoting the degradation of unfolded mutp53 protein, which identifies a new therapeutic opportunity for patients with TP53 mutations. Our results not only demonstrate triptolide-mediated inhibition of unfolded mutp53-induced NSCLC cell invasion and migration but also offer novel insights into the mechanisms underlying the therapeutic effects of triptolide. The specific degradation effect of triptolide on unfolded mutp53 proteins is expected to make triptolide a specific therapeutic drug for the treatment of NSCLC characterized by unfolded mutp53, providing early treatment for NSCLC patients with TP53 GOF mutation, thereby inhibiting NSCLC disease progression. TP53 mutation is an independent prognostic factor for NSCLC, and its specific treatment strategy will pave the way for effective individualized treatment of cancer patients.
Acknowledgments
The authors appreciate the academic support from the AME Lung Cancer Collaborative Group.
Funding: This work was financially supported by the China National Science Foundation (No. 81874396), Jinan Clinical Medicine Research Program for Thoracic Cancer (No. 201912007), and a grant from the Special Construction Project Fund for Taishan Mountain Scholars of Shandong Province.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-312/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-312/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-312/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All animal experiments were reviewed and approved by the Ethics Committee for Animal Experimentation of The Second Hospital of Shandong University (No. KYLL-2018(KJ)A-0046), in compliance with Institutional Animal Care and Use Committee guidelines. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sarode P, Mansouri S, Karger A, et al. Epithelial cell plasticity defines heterogeneity in lung cancer. Cell Signal 2020;65:109463. [Crossref] [PubMed]
- Chunhacha P, Chanvorachote P. Roles of caveolin-1 on anoikis resistance in non small cell lung cancer. Int J Physiol Pathophysiol Pharmacol 2012;4:149-55. [PubMed]
- Chiang YT, Chien YC, Lin YH, et al. The Function of the Mutant p53-R175H in Cancer. Cancers (Basel) 2021;13:4088. [Crossref] [PubMed]
- Li YR, Li S, Ho CT, et al. Tangeretin derivative, 5-acetyloxy-6,7,8,4'-tetramethoxyflavone induces G2/M arrest, apoptosis and autophagy in human non-small cell lung cancer cells in vitro and in vivo. Cancer Biol Ther 2016;17:48-64. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Xu F, Lin H, He P, et al. A TP53-associated gene signature for prediction of prognosis and therapeutic responses in lung squamous cell carcinoma. Oncoimmunology 2020;9:1731943. [Crossref] [PubMed]
- Liang S, Ezerskyte M, Wang J, et al. Transcriptional mutagenesis dramatically alters genome-wide p53 transactivation landscape. Sci Rep 2020;10:13513. [Crossref] [PubMed]
- Lang GA, Iwakuma T, Suh YA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 2004;119:861-72. [Crossref] [PubMed]
- Kalo E, Kogan-Sakin I, Solomon H, et al. Mutant p53R273H attenuates the expression of phase 2 detoxifying enzymes and promotes the survival of cells with high levels of reactive oxygen species. J Cell Sci 2012;125:5578-86. [Crossref] [PubMed]
- Eriksson M, Ambroise G, Ouchida AT, et al. Effect of Mutant p53 Proteins on Glycolysis and Mitochondrial Metabolism. Mol Cell Biol 2017;37:e00328-17. [Crossref] [PubMed]
- Sun S, Chen H, Sun L, et al. Hotspot mutant p53-R273H inhibits KLF6 expression to promote cell migration and tumor metastasis. Cell Death Dis 2020;11:595. [Crossref] [PubMed]
- Schulz-Heddergott R, Moll UM. Gain-of-Function (GOF) Mutant p53 as Actionable Therapeutic Target. Cancers (Basel) 2018;10:188. [Crossref] [PubMed]
- Liu J, Liu S, Wu Y, et al. Curcumin doped zeolitic imidazolate framework nanoplatforms as multifunctional nanocarriers for tumor chemo/immunotherapy. Biomater Sci 2022;10:2384-93. [Crossref] [PubMed]
- Nandi D, Cheema PS, Singal A, et al. Artemisinin Mediates Its Tumor-Suppressive Activity in Hepatocellular Carcinoma Through Targeted Inhibition of FoxM1. Front Oncol 2021;11:751271. [Crossref] [PubMed]
- Vendrely V, Amintas S, Noel C, et al. Combination treatment of resveratrol and capsaicin radiosensitizes pancreatic tumor cells by unbalancing DNA repair response to radiotherapy towards cell death. Cancer Lett 2019;451:1-10. [Crossref] [PubMed]
- Tian Z, Luo Y, Zhu J, et al. Transcriptionally elevation of miR-494 by new ChlA-F compound via a HuR/JunB axis inhibits human bladder cancer cell invasion. Biochim Biophys Acta Gene Regul Mech 2019;1862:822-33. [Crossref] [PubMed]
- Kupchan SM, Court WA, Dailey RG Jr, et al. Triptolide and tripdiolide, novel antileukemic diterpenoid triepoxides from Tripterygium wilfordii. J Am Chem Soc 1972;94:7194-5. [Crossref] [PubMed]
- Shao H, Ma J, Guo T, et al. Triptolide induces apoptosis of breast cancer cells via a mechanism associated with the Wnt/β-catenin signaling pathway. Exp Ther Med 2014;8:505-8. [Crossref] [PubMed]
- Yuan S, Wang L, Chen X, et al. Triptolide inhibits the migration and invasion of human prostate cancer cells via Caveolin-1/CD147/MMPs pathway. Biomed Pharmacother 2016;84:1776-82. [Crossref] [PubMed]
- Chen Z, Sangwan V, Banerjee S, et al. Triptolide sensitizes pancreatic cancer cells to TRAIL-induced activation of the death receptor pathway. Cancer Lett 2014;348:156-66. [Crossref] [PubMed]
- Li CJ, Chu CY, Huang LH, et al. Synergistic anticancer activity of triptolide combined with cisplatin enhances apoptosis in gastric cancer in vitro and in vivo. Cancer Lett 2012;319:203-13. [Crossref] [PubMed]
- Oliveira A, Beyer G, Chugh R, et al. Triptolide abrogates growth of colon cancer and induces cell cycle arrest by inhibiting transcriptional activation of E2F. Lab Invest 2015;95:648-59. [Crossref] [PubMed]
- Kaiser AM, Attardi LD. Deconstructing networks of p53-mediated tumor suppression in vivo. Cell Death Differ 2018;25:93-103. [Crossref] [PubMed]
- Chen M, Wang JM, Wang D, et al. Triptolide inhibits migration and proliferation of fibroblasts from ileocolonic anastomosis of patients with Crohn's disease via regulating the miR-16-1/HSP70 pathway. Mol Med Rep 2019;19:4841-51. [Crossref] [PubMed]
- Zhang W, Kang M, Zhang T, et al. Triptolide Combined with Radiotherapy for the Treatment of Nasopharyngeal Carcinoma via NF-κB-Related Mechanism. Int J Mol Sci 2016;17:2139. [Crossref]
- Bullock AN, Fersht AR. Rescuing the function of mutant p53. Nat Rev Cancer 2001;1:68-76. [Crossref] [PubMed]
- Baugh EH, Ke H, Levine AJ, et al. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ 2018;25:154-60. [Crossref] [PubMed]
- Pitolli C, Wang Y, Mancini M, et al. Do Mutations Turn p53 into an Oncogene? Int J Mol Sci 2019;20:6241. [Crossref] [PubMed]
- Zhu G, Pan C, Bei JX, et al. Mutant p53 in Cancer Progression and Targeted Therapies. Front Oncol 2020;10:595187. [Crossref] [PubMed]
- Muller P, Hrstka R, Coomber D, et al. Chaperone-dependent stabilization and degradation of p53 mutants. Oncogene 2008;27:3371-83. [Crossref] [PubMed]
- Tian Y, Li P, Xiao Z, et al. Triptolide inhibits epithelial-mesenchymal transition phenotype through the p70S6k/GSK3/β-catenin signaling pathway in taxol-resistant human lung adenocarcinoma. Transl Lung Cancer Res 2021;10:1007-19. [Crossref] [PubMed]
- Chaikovsky AC, Li C, Jeng EE, et al. The AMBRA1 E3 ligase adaptor regulates the stability of cyclin D. Nature 2021;592:794-8. [Crossref] [PubMed]
- The CLIP1-LTK Fusion Is an Oncogenic Driver of NSCLC. Cancer Discov 2022;12:286. [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Selkirk JK, Merrick BA, Stackhouse BL, et al. Multiple p53 protein isoforms and formation of oligomeric complexes with heat shock proteins Hsp70 and Hsp90 in the human mammary tumor, T47D, cell line. Appl Theor Electrophor 1994;4:11-8. [PubMed]
- Nagata Y, Anan T, Yoshida T, et al. The stabilization mechanism of mutant-type p53 by impaired ubiquitination: the loss of wild-type p53 function and the hsp90 association. Oncogene 1999;18:6037-49. [Crossref] [PubMed]
- Peng Y, Chen L, Li C, et al. Inhibition of MDM2 by hsp90 contributes to mutant p53 stabilization. J Biol Chem 2001;276:40583-90. [Crossref] [PubMed]
- Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014;505:495-501. [Crossref] [PubMed]
- Saleh MM, Scheffler M, Merkelbach-Bruse S, et al. Comprehensive Analysis of TP53 and KEAP1 Mutations and Their Impact on Survival in Localized- and Advanced-Stage NSCLC. J Thorac Oncol 2022;17:76-88. [Crossref] [PubMed]
- Labbé C, Cabanero M, Korpanty GJ, et al. Prognostic and predictive effects of TP53 co-mutation in patients with EGFR-mutated non-small cell lung cancer (NSCLC). Lung Cancer 2017;111:23-9. [Crossref] [PubMed]
- Vokes NI, Chambers E, Nguyen T, et al. Concurrent TP53 Mutations Facilitate Resistance Evolution in EGFR-Mutant Lung Adenocarcinoma. J Thorac Oncol 2022; Epub ahead of print. [Crossref] [PubMed]
- Mancini M, Righetto M, Noessner E. Checkpoint Inhibition in Bladder Cancer: Clinical Expectations, Current Evidence, and Proposal of Future Strategies Based on a Tumor-Specific Immunobiological Approach. Cancers (Basel) 2021;13:6016. [Crossref] [PubMed]
- Assoun S, Theou-Anton N, Nguenang M, et al. Association of TP53 mutations with response and longer survival under immune checkpoint inhibitors in advanced non-small-cell lung cancer. Lung Cancer 2019;132:65-71. [Crossref] [PubMed]
- Dhieb D, Belguith I, Capelli L, et al. Analysis of Genetic Alterations in Tunisian Patients with Lung Adenocarcinoma. Cells 2019;8:514. [Crossref] [PubMed]
- Ng JW, Lama D, Lukman S, et al. R248Q mutation--Beyond p53-DNA binding. Proteins 2015;83:2240-50. [Crossref] [PubMed]
- Boettcher S, Miller PG, Sharma R, et al. A dominant-negative effect drives selection of TP53 missense mutations in myeloid malignancies. Science 2019;365:599-604. [Crossref] [PubMed]
- Lorenzon I, Pellarin I, Pellizzari I, et al. Identification and Characterization of a New Platinum-Induced TP53 Mutation in MDAH Ovarian Cancer Cells. Cells 2019;9:36. [Crossref] [PubMed]
- Zhao Y, Li Y, Sheng J, et al. P53-R273H mutation enhances colorectal cancer stemness through regulating specific lncRNAs. J Exp Clin Cancer Res 2019;38:379. [Crossref] [PubMed]
- Butera G, Brandi J, Cavallini C, et al. The Mutant p53-Driven Secretome Has Oncogenic Functions in Pancreatic Ductal Adenocarcinoma Cells. Biomolecules 2020;10:884. [Crossref] [PubMed]
- Guo AK, Itahana Y, Seshachalam VP, et al. Mutant TP53 interacts with BCAR1 to contribute to cancer cell invasion. Br J Cancer 2021;124:299-312. [Crossref] [PubMed]
- Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu Rev Biochem 2008;77:557-82. [Crossref] [PubMed]
- Lambert JM, Moshfegh A, Hainaut P, et al. Mutant p53 reactivation by PRIMA-1MET induces multiple signaling pathways converging on apoptosis. Oncogene 2010;29:1329-38. [Crossref] [PubMed]
- Yu X, Vazquez A, Levine AJ, et al. Allele-specific p53 mutant reactivation. Cancer Cell 2012;21:614-25. [Crossref] [PubMed]
- Hiraki M, Hwang SY, Cao S, et al. Small-Molecule Reactivation of Mutant p53 to Wild-Type-like p53 through the p53-Hsp40 Regulatory Axis. Chem Biol 2015;22:1206-16. [Crossref] [PubMed]
- Jiang Y, Rossi P, Kalodimos CG. Structural basis for client recognition and activity of Hsp40 chaperones. Science 2019;365:1313-9. [Crossref] [PubMed]
- Song YJ, Zhong CB, Wang XB. Heat shock protein 70: A promising therapeutic target for myocardial ischemia-reperfusion injury. J Cell Physiol 2019;234:1190-207. [Crossref] [PubMed]


