
Longitudinal prediction of lung nodule invasiveness by sequential modelling with common clinical computed tomography (CT) measurements: a prediction accuracy study
Introduction
Lung cancer is the leading cause of death among all malignancies, with a 5-year survival rate of no more than 20% (1). National prospective clinical trials have confirmed that lung cancer screening by low-dose computed tomography (LDCT) can reduce the mortality rate by 20% compared with chest X-rays (2). Lung nodules are the early radiological signs of lung cancer on LDCT scans. Numerous radiological societies have established guidelines for managing these nodules on the basis of their characteristics at baseline, and more importantly, on their changing attributes on follow-up scans (3-5). Longitudinal imaging profiling of pulmonary nodules can identify fast growing nodules with a high suspicion of being malignancy requiring further management, which involves invasive procedures such as biopsy or surgical resection. However, fast growth is only indicative, not conclusive, and post-operative pathological diagnosis may deem the invasive procedures unnecessary. Lung adenocarcinoma is the most common type of lung cancer, and usually presents as pure ground-glass nodules (pGGN) or subsolid nodules on CT scans. Less invasive adenocarcinomas, including atypical adenomatous hyperplasia (AAH), adenocarcinoma in situ (AIS), and minimally invasive adenocarcinoma (MIA), have an excellent prognosis with surveillance alone, while invasive adenocarcinoma (IAC) has a poor prognosis, and is better managed by surgical resection (6). Therefore, to facilitate optimal management, pre-operative determination of both the growth patterns of pulmonary nodules and their invasiveness is desirable.
Various approaches for the preoperative imaging-based prediction of nodule invasiveness on CT scans have shown promising results, including conventional machine learning, radiomics, and more recently, deep learning (7-9). Both conventional machine learning and radiomics try to map manually defined features to classification targets using classifiers including logistic regression (LR) and random forests, and differ in the inputs that they accept. The inputs to the conventional machine learning are usually semantic features that are limited in number, but can be interpreted by experts and are commonly used by clinicians to characterize lung nodules. Examples of these features include size, attenuation pattern, and lobulation (10). The inputs to radiomics methods consist of hundreds of radiomic features, which are much less intuitive and interpretable, and are extracted from three-dimensional regions of nodules on CT images using algorithms based on predefined mathematical formulae (7,8). Deep learning, which is built on multiple stacked layers of neural networks, can map raw data signals (from CT images in our case) directly to targets and learn discriminative features during the supervised training process, without requiring human input to direct this learning process (11). As in other domains, deep learning has achieved remarkable performance in medical imaging analysis, and has reached a level of performance close to that of radiologists in discerning the lung nodule phenotypes (9). Despite their success, these efforts remain suboptimal, as they only focus on a ‘snapshot’ of a nodule, neglecting the rich longitudinal data that clinicians routinely acquire during follow-up, and which can track growth patterns. Lung nodules may change morphologically over time via interaction with the human immune system, and a snapshot may be insufficient to capture the necessary information and make accurate predictions.
The recurrent neural network (RNN) is a well-known sequential modelling method that can handle longitudinal data. First proposed in 1997, the long short-term memory (LSTM) network (12) is a go-to RNN that is being increasingly adopted for sequential modelling in multiple fields, including natural language processing, image captioning, and medical imaging (13,14). Numerous classification studies have demonstrated that sequential modelling accepting longitudinal or time-series data as inputs outcompeted cross-sectional modelling that only received data from a single time point. These classification tasks include abnormalities detection on chest X-ray (15), focal liver lesion classification (16), identification of coronavirus disease 2019 (COVID-19) patients prone to malignant progression (17), and prediction of nodule malignancy on CT (18). Sequential modelling using an LTSM has also been used to predict the invasiveness of pulmonary nodules using consecutive serial CT images (19). However, this study only included CT images from two consecutive time points as inputs, excluding the remaining longitudinal data that is commonly available for patients with lung nodules. Therefore, the full potential of sequential modelling has not yet been realized, at least in terms of predicting tumor invasiveness.
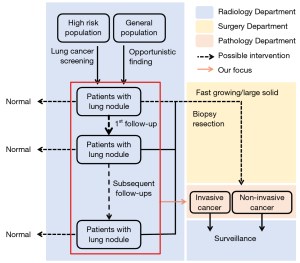
In this study, we aimed to harness the sequential modelling abilities of an LSTM to build a high-performance classifier, in order to pre-operatively predict the invasiveness of lung nodules using semantic features that are commonly used in clinics and were taken from all historical examinations available for each individual patient (Figure 1). The patients could differ in the number of CT examinations they underwent for their lung nodules, as well as the intervals between any two consecutive CT examinations. We chose semantic features (mainly size and intensity) for the modelling because they are easy to obtain and are routinely used by clinicians to assess growth patterns with decent accuracy. In contrast, radiomic features often depend on the delineation of nodule contours, while deep learning requires the registration of multiple series of CT scans to locate the same nodule. We explored ways to optimize the LSTM-based classifier by tuning the model hyper-parameters and experimenting on different combinations of time-invariant and time-varying nodule features. To showcase the superiority of sequential modelling by LSTM, we also built LR classifiers for comparison purposes. We present the following article in accordance with the STARD reporting checklist (Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-319/rc).
Methods
Data collection
This prediction accuracy study was approved by the Institutional Review Board of Shanghai Chest Hospital (No. KS1956). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Given the retrospective nature of the analysis, the Board waived the requirement for patients’ written consent, and anonymity was ensured for all patient data.
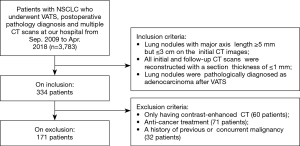
We retrospectively reviewed the records of patients who were found to have pulmonary subsolid nodules on CT scans, were followed-up at least once, and finally underwent video-assisted thoracoscopic surgery (VATS) together with postoperative pathological diagnosis of nodules at Shanghai Chest Hospital between September 2009 and April 2018. The inclusion criteria for this study were as follows: (I) patients’ lung nodules should have a major-axis length of ≥5 mm but ≤3 cm on the initial CT scan; (II) the initial and follow-up CT images of the patients should have a section thickness ≤1 mm; and (III) the postoperative pathological diagnosis of the nodules should be lung adenocarcinoma. The exclusion criteria were as follows: (I) patients who received any anti-cancer treatment such as chemotherapy and targeted therapy during follow-up; (II) patients who only received a contrast-enhanced scan during any CT examination visit; and (III) patients with a history of previous or concurrent malignancy. A flow chart of the inclusion and exclusion process is shown in Figure 2.
The pathological diagnosis was performed by two experienced pathologists according to the 2015 World Health Organization (WHO) classification of lung tumours. On the basis of the presence of invasive components and the degree of invasion, the samples were divided into two groups. The first group included AIS (with no invasive features) and MIA (≤5 mm invasion), and the second group was IA (>5 mm invasion).
Image acquisition
The included patients were scanned with multi-detector CT scanners (Brilliance 64, Ingenuity CT128, Brilliance iCT256; Philips Medical Systems, Cleveland, OH, USA) using the following CT parameters: tube voltage, 120 kVp; tube current, 250 mA; pitch, 0.984; and thickness, 1 mm. CT images were reconstructed at 1.0 mm thickness and 1 mm interval using a sharp reconstruction kernel (C filter).
Modelling
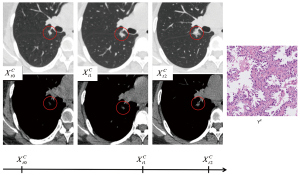
To characterize the nodules in the study, two pulmonary radiologists with over 5 years of experience identified the morphological types of the nodules and measured their size and intensity on the CT scans of each examination using ITK-SNAP software (version 3.8.0; http://www.itksnap.org), according to the recommendations of the Fleischner Society (20). The longitudinal data of a typical patient in our study are presented in Figure 3. The morphological types were solid, part solid, and GGN. For the size of the nodules, the major axis length (Lmajor) and minor axis length (Lminor) on the axial view were obtained on both the lung window (LW; level, −520 HU; width, 1,450 HU) and mediastinal window (MW) settings (level, 50 HU; width, 400 HU). For intensity, we roughly drew a sphere-like region of interest of the nodules and then recorded the mean CT value (I) inside it. We approximated the volume (V) of the nodules at a particular time point based on the Lmajor and Lminor on the lung window settings according to the following formula:
For each nodule, we also computed the time interval (T) in days since the initial discovery for every examination visit. The time interval at baseline examination was taken as zero. Using the Vinital at baseline examination, Vfinal at preoperative examination, and the duration between the two time points (T2ends), we computed the volume doubling time (21) according to the following formula:
In total, we obtained seven time-specific features for a nodule at a particular time point, including four lengths consisting of Lmajor and Lminor on both LW and MW, intensity (I) of the mean CT value, time interval (T) since initial discovery, and approximate volume (V). The seven time-varying features, together with the four time-invariant features of nodule location, morphology, patient age, and gender, formed a pool of semantic features used to build the classifiers for distinguishing less invasive nodules (IAC) (AAH, AIS, and MIA) from IAC.
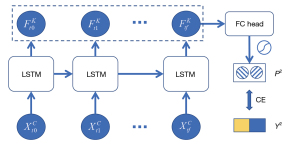
To provide baseline models for comparison, we developed three LR-based classifiers using nodule features at baseline, pre-surgery, and the two-ends of the follow-up sequence. To sequentially model the longitudinal data, we trained an LSTM-based classifier using nodule features measured on two or more examinations available to each patient. An architectural overview of the LSTM classifier is depicted in Figure 4. The LSTM classifier consisted of one fully connected (FC) layer, followed by a batch normalization layer, three LSTM layers, and finally a projection head outputting two scores as prediction. We applied the following equations iteratively from t=1 to T to update the hidden states:
where i, o, f, and c are the input gate, output gate, forget gate, and cell activation vectors, respectively; σ is the sigmoid function; g is the cell input activation function; and h is the cell output activation function. Wf, Wi, Wo, Wc denote the weight matrices, and bf, bi, bo, bc denote the bias vectors.
Since the seven nodule features differed in scale, we normalized them separately using their individual mean and standard deviation over all of the training samples, so as to achieve better generalization.
Statistical analysis
We also conducted chi-square tests of independence to examine the relationship between nodule invasiveness and other categorical variables, including nodule location, nodule morphology, number of CT scans, and the patient’s gender. For the seven time-specific features, we performed independent t-tests to evaluate the differences between the IAC group and the less-IAC group. With limited data samples, we applied five-fold cross validation to test the performance of the proposed classifiers, which is considered more robust than a single split of the whole dataset into training and testing set. The area under the curve (AUC) of the receiver operating characteristics (ROC) curve is used as the main indicator of the performance of all of the classifiers. The Delong’s test was employed to deciding if the difference of AUC between models reaching statistical significance (22). All open-source packages modelling and statistical analyses were run in Python 3.8 using open-source packages PyTorch (v1.7.1 + cu110) (23), Pytorch-lightning (v1.3.8), Scikit-learn (v0.24.2), and Researchpy (v0.3.2).
Results
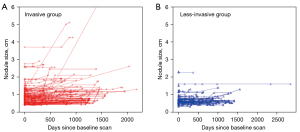
In total, 171 patients were finally included, consisting of 58 men and 113 women with a mean age of 59.04±10.17 years. Each patient had one valid lung nodule for analysis. All nodules were followed up at least once over an average of 784.64±537.44 days. At baseline, the mean sizes of the nodules were 1.03±0.62 cm in the major axis and 0.80±0.41 cm in the minor axis, with an intensity of −428.41±249.94 HU. Postoperative biopsy identified 59 less-invasive cancers and 112 invasive adenocarcinomas. These two invasiveness groups differed significantly in terms of gender, age, nodule morphology, nodule size, and intensity (see Table 1). As shown in Figure 5, the temporal changes in the sizes of individual nodules across all CT examination visits are presented separately for the less-invasive and invasive groups.
Table 1
| Characteristics | Less invasive cancers | Invasive cancers | Statistics (𝜒2/t test) |
|---|---|---|---|
| Gender (N) | 9.38** | ||
| Male | 11 | 47 | |
| Female | 48 | 65 | |
| Age (years) | 56.14±12.24 | 60.56±8.56 | −2.76** |
| Nodule location | 1.71 | ||
| Left upper | 14 | 33 | |
| Left lower | 7 | 10 | |
| Right upper | 26 | 42 | |
| Right middle | 3 | 9 | |
| Right lower | 9 | 12 | |
| Nodule morphology | 15.70** | ||
| Solid | 1 | 20 | |
| Part-solid | 0 | 9 | |
| Pure ground-glass | 58 | 83 | |
| Nodule feature on baseline | |||
| Lmajor on lung window (cm) | 0.84±0.44 | 1.13±0.67 | −3.07** |
| Lminor on lung window (cm) | 0.70±0.35 | 0.85±0.43 | −2.22* |
| Lmajor on mediastinal window (cm) | 0.01±0.08 | 0.28±0.68 | −3.07** |
| Lminor on mediastinal window (cm) | 0.01±0.06 | 0.20±0.45 | −3.18** |
| V on lung window (cm3) | 0.47±1.13 | 1.07±2.90 | −1.52 |
| I (mean CT value in manual ROI) | −506.43±169.04 | −387.30±275.33 | −3.03** |
| Volume doubling time (years Approx.) | 41.54±57.32 | 21.71±45.46 | 2.47* |
| Duration of follow-ups (days) | 703.88±556.66 | 827.18±524.56 | −1.43 |
| Number of follow-ups (N) | 4.39 | ||
| 1 | 1 | 1 | |
| 2 | 14 | 16 | |
| 3 | 6 | 20 | |
| 4 | 13 | 21 | |
| 5 | 7 | 18 | |
| 6 | 18 | 36 |
*, P<0.05; **, P<0.01. Lmajor and Lminor are the nodule lengths in the major axis and minor axis on the axial view. CT, computed tomography; ROI, region of interest. Values are mean ± standard deviation.
When training the conventional LR for binary classification of nodule invasiveness, we attempted to use all 11 nodule features as input variables, but found that volume measured using the lung window setting (Vlw) failed to fit the model, and therefore, we only used the other 10 variables in the modelling. In the cross-validation, the LR model built using nodule variables at baseline (LR_N10@Baseline) performed significantly worse than the LR model built using variables at preoperative CT scan (LR_N10@Final) (AUC 0.716 vs. 0.885, respectively, P<0.001). The LR_N10@Final model in turn performed significantly worse than the LR built using features from both time points (LR_N10@2Ends; 0.885 vs. 0.947, respectively, P<0.001) (see Figure 6A).
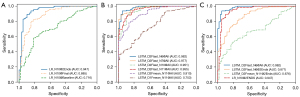
To optimize the LSTM-based classifier, we evaluated several hyper-parameters, including the directionality of LSTM layers (D1 or D21), and experimented with how to use the LSTM embedded feature vectors from a mutable number of time points for classification (Fmean or Flast2), which data points to include in the modelling (the baseline and pre-surgery visits or all visits), and which nodule variables to use for input to the models (11, 10, 7, or 63). The performance measures of all LSTM-based classifiers experimented with are summarized in Table 2. Nodule variables at the baseline and preoperative visits did not produce a high-performing LSTM-based classifier (LSTM_D2Fmean_N11@2End), with an AUC of only 0.676, which was significantly worse than the LR-based classifier (LR_N10@2Ends). Therefore, we used nodule variables from all examination timepoints in the remaining LSTM models. Within the LSTM models, the bidirectional LSTM performed better than the unidirectional LSTM (AUCs: 0.910 versus 0.753), and Flast performed better than Fmean (AUCs: 0.955 versus 0.910). Four unchanging nodule features, including nodule location, nodule morphology, as well as the patient’s gender and age, were unhelpful in the LSTM models (0.955 versus 0.977 for LSTM_D2Flast_N11@All and LSTM_D2Flast_N7@All, respectively). The LSTM-based classifier worked slightly better without Vlw as an input variable (0.977 versus 0.982 for LSTM_D2Flast_N7@All and LSTM_D2Flast_N6@All, respectively). The ROC curves of these LSTM models are shown in Figure 6B.
Table 2
| Classifiers | Input variables | AUC (95% CI)d | Accuracy | Sensitivity | Specificity | |
|---|---|---|---|---|---|---|
| Number of variablesc | Timepoint(s) | |||||
| Logistic regression | 11 | Baseline | Failed to fit | |||
| 10 | Baseline | 0.716 (0.636–0.795)* | 0.678 | 0.804 | 0.441 | |
| 10 | Pre-surgery | 0.885 (0.831–0.939)* | 0.807 | 0.848 | 0.729 | |
| 10 | Two ends | 0.947 (0.908–0.986)* | 0.906 | 0.938 | 0.847 | |
| LSTM | ||||||
| D1a; Fmeanb | 11 | Two ends | 0.676 (0.595–0.756)* | 0.585 | 0.688 | 0.390 |
| D1; Fmean | 11 | All visits | 0.753 (0.680–0.826)* | 0.673 | 0.759 | 0.508 |
| D2; Fmean | 11 | All visits | 0.910 (0.868–0.951)* | 0.825 | 0.893 | 0.695 |
| D2; Flast | 11 | All visits | 0.955 (0.927–0.982)* | 0.871 | 0.893 | 0.830 |
| D2; Flast | 10 | All visits | 0.951 (0.922–0.981)* | 0.883 | 0.929 | 0.780 |
| D2; Flast | 7 | All visits | 0.977 (0.961–0.994)* | 0.900 | 0.920 | 0.864 |
| D2; Flast | 6 | All visits | 0.982 (0.966–0.997)* | 0.924 | 0.946 | 0.881 |
a, D1 stands for unidirectional LSTM; D2 stands for bidirectional LSTM; b, means that the embedding features to the LSTM from all time points were averaged first and then sent to the FC head, while Flast indicates that only the embedding features from the last time point were sent to the FC head for classification; c, each nodule was characterized by the 11 variables specified in the Methods section. Using logistic regression, only 10 variables were found to fit the model, and the volume on lung window setting (Vlw) was excluded. For the LSTM, we experimented with four combinations of variables: all 11 variables, 10 variables with Vlw removed, seven with the four unchanging ones (nodule location, nodule morphology, patient gender, and patient age) removed, and 6 variables with Vlw also removed from the set of seven variables; d, Delong’s test was used to compare the AUC (95% CI) performance of all classifiers with that of the logistic regression using 10 feature inputs from both the baseline visit and the preoperative visit (LR_N10@2Ends). Statistically significant differences were labeled as: *, P<0.001. AUC, area under the curve; CI, confidence interval; LSTM, long short-term memory.
Finally, the best LSTM model (LSTM_D2Flast_N6@All) surpassed the best LR model (LR_N10@2Ends) in the binary classification task of nodule invasiveness, and the difference was statistically significant (0.982 versus 0.947, P<0.05). The ROC curves of both models are shown in Figure 6C.
Discussion
Preoperative prediction of the invasiveness of lung nodules can avoid unnecessary invasive procedures for patients with a good prognosis, and has attracted research attention amid the excitement generated by advances in artificial intelligence. Most previous studies have attempted to make predictions based on data from a single time point, with the hope of minimizing the number of follow-ups as well as the associated radiation risk and economic burden (7,8). However, the goal of accurate prediction might not be so achievable, and the precision of such an approach cannot be guaranteed, with follow-ups being essential for clinicians to characterize the growth patterns of indeterminate lung nodules and recommend management strategies. A study has attempted to exploit follow-up data through sequential modelling for phenotyping lung nodules (21). Continuing this line of research, we conducted extensive experiments to push the performance limit using semantic features that are easily obtained in clinics and represent the entire imaging history of each individual patient. Through cross-validation, we found that a bidirectional LSTM with time-varying features from all available time points as input (LSTM_D2Flast_N6) performed best in predicting the invasiveness of lung nodules. Confirming our hypothesis, this optimal LSTM-based classifier also outperformed the best LR models, which did not possess such sequential modelling capacities. Through further verification using a larger dataset, we found that our LSTM system has the potential to be integrated into the clinical pipeline of nodule management, so as to reduce unnecessary invasive procedures and costs.
In this study, we selected a range of lung nodule features that are commonly measured in the clinic (mainly size and intensity measures) to build an imaging biomarker (24) for nodule invasiveness. As shown in Table 1, most of the features at baseline examination were associated with nodule invasiveness, as shown in previous studies (7,8). For instance, a study has reported that a rapid change in nodule intensity was associated with faster growth in lung nodules (25). Studies also demonstrated that the VDT of invasive lung adenocarcinoma (showing as a subsolid nodule on CT) was significantly shorter than that of its non-invasive counterpart (21,26). The high performance of the models built in this study supports the idea that these simple, interpretable, and easy-to-obtain imaging features are capable of building accurate classifiers for nodule phenotyping. Previous study has also shown that the semantic features of nodules are not inferior to radiomic features for building accurate classifiers of nodule malignancy (27).
The use of these simple features also alleviates the challenge of sequential modelling of irregular longitudinal data, which occurs because the follow-up for lung nodules is often not regularly spaced, and the total number of examinations can vary from patient to patient (Figure 3). A naive LSTM network expects regularly sampled data and does not consider the varying interval between data points (15). Our study treated the time elapse of each follow-up since baseline examination as a nodule feature that could be directly input into the LSTM, thereby achieving promising results. This intuitive approach was feasible in our study because we only used a small number of semantic features at any particular time point, and the interval feature could therefore exert a significant influence. To the contrary, previous studies on sequential modelling used high-dimensional discriminative features, usually involving hundreds of dimensions, extracting them using a convolutional neural network, and a single interval feature could easily be overwhelmed (15,18). The solution offered in previous studies was to incorporate the time into the computational process of the LSTM as weighting factors for the high-dimensional feature vector. Although less intuitive, such methods also have merit, and are worthy of investigation in future studies (15,18).
Our study also demonstrated the superiority of using the entire imaging history available for each patient (LSTM_D2Flast_N6@All) over only using data from a fixed number of timepoints (LSTM_D2Flast_N6@2Ends). This finding is consistent with a previous treatment response study that also used serial CT data (28), and another deep learning study that predicted the future contours of the tumor after radiation therapy (14). Xu et al. explored the same task of preoperative determination of nodule invasiveness as the present study, and only used CT scans at two time points as inputs into their sequential model, without considering the interval in between them (19), which might have contributed to their suboptimal performance. Similarly, Gao et al. built a Convolutional Neural Networks (CNN)-LSTM hybrid sequential model to predict the likelihood of nodule malignancy and explored ways to model temporal dependency for both regular and irregular longitudinal data (18). However, they only used two time points in their modelling, fixed and limited. Sacrificing parts of the longitudinal data of some patients to accommodate others with less data points might have resulted in the less promising performance of their model. Again, our method considered all of the longitudinal data available for each patient and thus achieved higher accuracy.
Follow-up data are essential for managing lung nodules found on CT scans, in terms of both guidelines and in clinical practice. Our study provides further empirical evidence illustrating that such sequential lung nodule data can be modelled by LSTMs for classification purposes, and we were able to improve on the performance of a conventional LR classifier by a significant margin (AUC 0.982 versus 0.947). To make the comparison rigorous, we developed three baseline models using LR. Within these baseline models, some interesting observations were made. First, the preoperative LR (LR_N10@Final) performed better than the baseline LR (LR_N10@baseline), which made sense because the preoperative examination was temporally closer to the nodule’s pathological diagnosis compared to the baseline examination, and the cellular composition of the nodules would evolve over time. Digumarthy et al. also found that radiomic features measured from lung nodules on preoperative CT were more predictive for nodule malignancy than the same radiomic features measured on baseline CT (29). Second, the LR_N10@2Ends model, which combined both baseline and preoperative CT image features, achieved the highest accuracy among the three LR models, which suggests that although LR is incapable of sequential modelling, it could benefit from additional information from more time points.
When optimizing the LSTM-based classifiers, we made some interesting findings that may be helpful for futures studies on sequential modelling. The last embedded feature was more discriminative than the averaged embedded features from all time points. Also, the exclusion of time invariant features, namely nodule location, nodule morphology, patient gender, and patient age, also boosted performance. The estimated VDT did not contribute to the model performance.
Our study has several limitations that should be noted. First, we did not use external data to evaluate our models, which may have hindered the generalizability of our method. In this study, we only evaluated the performance of our model on positive samples with confirmed early-stage lung cancer; thus, the behavior of our model on negative samples is unknown and should be investigated in future studies. Secondly, the semantic features used in our study were obtained by radiologists and may have been influenced by subjectivity and individual bias, and may therefore suffer from interobserver variability. Despite being diagnostic, these features were limited in number and might not have fully captured characteristics that are useful for discerning nodule invasiveness. Future studies can explore ways to integrate more features into the LSTM system, including radiomic features and CNN-extracted features, as reported by Fang et al. (17). Thirdly, some nodules in our study that were confirmed as early stage lung cancer did not increase in size or grew very slowly over a long period, as shown in Figure 5, and therefore deviated from the exponential growth pattern described in previous studies (26,30). Since the focus of our study was on the sequential modelling, we did not make efforts to analyze these nodules using indolent growth patterns, although this is worth exploring in future studies.
Acknowledgments
The authors appreciate the academic support from the AME Radiology Collaborative Group.
Funding: This work was financially supported by the National Natural Science Foundation of China (Nos. 81871353; 82071873); the Shanghai Municipal Health Commission Project (Nos. 2019SY063; 20204Y0201); the Shanghai Key Laboratory Open Project (No. STCSM18DZ2270700); the Shanghai “Rising Stars of Medical Talents” Youth Development Program [No. SHWSRS(2021)_099]; and the China International Medical Foundation (No. Z-2014-07-2003-20).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-319/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-319/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-319/coif). DS is an employee of Keya Medical Technology Co. Ltd. The company provided the computer with GPU cards that were used to conduct the model training, testing and data analysis for this study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This prediction accuracy study was approved by the Institutional Review Board of Shanghai Chest Hospital (No. KS1956). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Given the retrospective nature of the analysis, the Board waived the requirement for patients’ written consent, and anonymity was ensured for all patient data.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
1, D1 indicates unidirectional LSTM and D2 indicates bidirectional.
2, Fmean represents the model in which the LSTM-embedded features of all time points are averaged first and then sent to the FC head, whereas Flast indicates that only the LSTM-embedded features from the last time point were sent to the FC head for classification.
3, 11 represents all variables available in this study; 10 includes all variables except for the volume on lung window setting (Vlw); 7 means that four variables unchanging across follow-up (nodule location, nodule morphology, patient gender, and patient age) were discarded; 6 signifies that Vlw was discarded from the set of 7.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Oudkerk M, Liu S, Heuvelmans MA, et al. Lung cancer LDCT screening and mortality reduction - evidence, pitfalls and future perspectives. Nat Rev Clin Oncol 2021;18:135-51. [Crossref] [PubMed]
- Baldwin DR, Callister MEGuideline Development Group. The British Thoracic Society guidelines on the investigation and management of pulmonary nodules. Thorax 2015;70:794-8. [Crossref] [PubMed]
- MacMahon H, Naidich DP, Goo JM, et al. Guidelines for Management of Incidental Pulmonary Nodules Detected on CT Images: From the Fleischner Society 2017. Radiology 2017;284:228-43. [Crossref] [PubMed]
- Mazzone PJ, Silvestri GA, Patel S, et al. Screening for Lung Cancer: CHEST Guideline and Expert Panel Report. Chest 2018;153:954-85. [Crossref] [PubMed]
- Yanagawa N, Shiono S, Abiko M, et al. New IASLC/ATS/ERS classification and invasive tumor size are predictive of disease recurrence in stage I lung adenocarcinoma. J Thorac Oncol 2013;8:612-8. [Crossref] [PubMed]
- Fan L, Fang M, Li Z, et al. Radiomics signature: a biomarker for the preoperative discrimination of lung invasive adenocarcinoma manifesting as a ground-glass nodule. Eur Radiol 2019;29:889-97. [Crossref] [PubMed]
- Tao G, Yin L, Shi D, et al. Dependence of radiomic features on pixel size affects the diagnostic performance of radiomic signature for the invasiveness of pulmonary ground-glass nodule. Br J Radiol 2021;94:20200089. [Crossref] [PubMed]
- Gong J, Liu J, Hao W, et al. A deep residual learning network for predicting lung adenocarcinoma manifesting as ground-glass nodule on CT images. Eur Radiol 2020;30:1847-55. [Crossref] [PubMed]
- Liu Y, Wang H, Li Q, et al. Radiologic Features of Small Pulmonary Nodules and Lung Cancer Risk in the National Lung Screening Trial: A Nested Case-Control Study. Radiology 2018;286:298-306. [Crossref] [PubMed]
- Esteva A, Robicquet A, Ramsundar B, et al. A guide to deep learning in healthcare. Nat Med 2019;25:24-9. [Crossref] [PubMed]
- Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput 1997;9:1735-80. [Crossref] [PubMed]
- Bai W, Suzuki H, Qin C, et al. Recurrent Neural Networks for Aortic Image Sequence Segmentation with Sparse Annotations. In: Frangi AF, Schnabel JA, Davatzikos C, et al., editors. Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. Cham: Springer International Publishing, 2018:586-94.
- Wang C, Rimner A, Hu YC, et al. Toward predicting the evolution of lung tumors during radiotherapy observed on a longitudinal MR imaging study via a deep learning algorithm. Med Phys 2019;46:4699-707. [Crossref] [PubMed]
- Santeramo R, Withey S, Montana G. Longitudinal Detection of Radiological Abnormalities with Time-Modulated LSTM. In: Stoyanov D, Taylor Z, Carneiro G, et al., editors. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Cham: Springer International Publishing, 2018:326-33.
- Liang D, Lin L, Hu H, et al. Combining Convolutional and Recurrent Neural Networks for Classification of Focal Liver Lesions in Multi-phase CT Images. In: Frangi AF, Schnabel JA, Davatzikos C, et al., editors. Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. Cham: Springer International Publishing, 2018:666-75.
- Fang C, Bai S, Chen Q, et al. Deep learning for predicting COVID-19 malignant progression. Med Image Anal 2021;72:102096. [Crossref] [PubMed]
- Gao R, Tang Y, Xu K, et al. Time-distanced gates in long short-term memory networks. Med Image Anal 2020;65:101785. [Crossref] [PubMed]
- Xu Y, Li Y, Yin H, et al. Consecutive Serial Non-Contrast CT Scan-Based Deep Learning Model Facilitates the Prediction of Tumor Invasiveness of Ground-Glass Nodules. Front Oncol 2021;11:725599. [Crossref] [PubMed]
- Bankier AA, MacMahon H, Goo JM, et al. Recommendations for Measuring Pulmonary Nodules at CT: A Statement from the Fleischner Society. Radiology 2017;285:584-600. [Crossref] [PubMed]
- Heuvelmans MA, Oudkerk M, de Bock GH, et al. Optimisation of volume-doubling time cutoff for fast-growing lung nodules in CT lung cancer screening reduces false-positive referrals. Eur Radiol 2013;23:1836-45. [Crossref] [PubMed]
- Sun X, Xu W. Fast Implementation of DeLong’s Algorithm for Comparing the Areas Under Correlated Receiver Operating Characteristic Curves. IEEE Signal Process Lett 2014;21:1389-93. [Crossref]
- Paszke A, Gross S, Massa F, et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In: Advances in Neural Information Processing Systems. Curran Associates, Inc.; 2019. Available online: https://papers.nips.cc/paper/2019/hash/bdbca288fee7f92f2bfa9f7012727740-Abstract.html
- O'Connor JP, Aboagye EO, Adams JE, et al. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol 2017;14:169-86. [Crossref] [PubMed]
- Bak SH, Lee HY, Kim JH, et al. Quantitative CT Scanning Analysis of Pure Ground-Glass Opacity Nodules Predicts Further CT Scanning Change. Chest 2016;149:180-91. [Crossref] [PubMed]
- de Margerie-Mellon C, Ngo LH, Gill RR, et al. The Growth Rate of Subsolid Lung Adenocarcinoma Nodules at Chest CT. Radiology 2020;297:189-98. [Crossref] [PubMed]
- Hancock MC, Magnan JF. Lung nodule malignancy classification using only radiologist-quantified image features as inputs to statistical learning algorithms: probing the Lung Image Database Consortium dataset with two statistical learning methods. J Med Imaging (Bellingham) 2016;3:044504. [Crossref] [PubMed]
- Xu Y, Hosny A, Zeleznik R, et al. Deep Learning Predicts Lung Cancer Treatment Response from Serial Medical Imaging. Clin Cancer Res 2019;25:3266-75. [Crossref] [PubMed]
- Digumarthy SR, Padole AM, Rastogi S, et al. Predicting malignant potential of subsolid nodules: can radiomics preempt longitudinal follow up CT? Cancer Imaging 2019;19:36. [Crossref] [PubMed]
- Heuvelmans MA, Vliegenthart R, de Koning HJ, et al. Quantification of growth patterns of screen-detected lung cancers: The NELSON study. Lung Cancer 2017;108:48-54. [Crossref] [PubMed]


