
Re-evaluating the need for mediastinal lymph node dissection and exploring lncRNAs as biomarkers of N2 metastasis in T1 lung adenocarcinoma
Introduction
Lung cancer has the highest mortality rate of malignant tumor types worldwide (1). Non-small cell lung cancer (NSCLC) accounts for 85% of lung cancers, with more than half of these cases presenting histologically as adenocarcinoma (LUAD). Currently, surgery remains the mainstay of curative treatment, particularly for early-stage cancers. Mediastinal lymph node dissection (MLND) is a critical part of traditional curative lung cancer resection, providing benefits in accurate clinical staging and survival (2). However, MLND also may carry elevated risk of perioperative complications.
The frequency of diagnosing early-stage lung cancers has increased in recent years, potentially related to greater imaging technology and the rise of lung cancer screening. These early staged cases have led some investigators to the need for uniform MLND in all patients, given the lesser incidence of occult pathologic N2 metastasis and controversy over survival benefits in patients with earlier clinical stages (3). Indeed, the key challenge and ultimate goal both lie in improving the accuracy and reliability of preoperatively providing a clinical N status that parallels pathological N status (4). The clinical TNM stage is currently diagnosed by computed tomography (CT) and positron emission tomography combined with CT (PET/CT). However, CT and PET/CT have limitations in identifying N2 metastasis, with similar appearances to nodes that are hyperplastic or inflammatory. Invasive mediastinal nodal staging through mediastinoscopy and endobronchial ultrasound guided transbronchial needle aspiration (EBUS-TBNA) add substantially to the accuracy, though with need for additional interventions carrying their own risk profiles, costs, and inconveniences. Therefore, diagnostic biomarkers of N2 lymph node metastasis could be of substantial benefit in guiding clinical treatment, but which are deficient currently.
Long non-coding RNAs (lncRNAs) are a class of RNAs of more than 200 nucleotides in length that do not encode proteins. A previous study has shown that lncRNAs can modulate carcinogenesis and influence metastasis and invasion in various types of cancer (5). Consequently, several lncRNAs have the potential to be a biomarker for diagnosis, prognosis, and resistance for treatment in several cancers (6-12). LncRNAs may be effective biomarkers predicting N2 stage because of their expression specificity and powerful biological functions (13).
This study explored the benefit of MLND in clinical T1 NSCLC by comparing prognosis among patients who did not receive MLND and received MLND (subdivided in pN0–1 group and pN2 group). Subsequently, the correlation of pN2 metastasis with basic clinical characteristics was analyzed in pT1 NSCLC. Furthermore, as a preliminary search for biomarkers, we screened differentially expressed lncRNAs in pT1 LUAD, evaluated their diagnostic value to detect N2 metastasis, and further analyzed their prognostic significance. We present the following article in accordance with the STARD reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-207/rc).
Methods
Data collection
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). We selected those T1 stage NSCLC patients with complete clinical information and follow-up data, and who had undergone surgical resection (lobectomy or pneumonectomy) [2010–2015] from the Surveillance, Epidemiology, and End Results (SEER) database of the National Cancer Institute (NCI) (https://seer.cancer.gov/) (14) using SEER*Stat (8.3.9.2), to analyze the influence of MLND on postoperative prognoses. The SEER program registries routinely collect demographic and clinic data on patients, and the mortality data reported by SEER are provided by the National Center for Health Statistics. Next, the HTseq-count and corresponding clinical pathologic information of T1 stage NSCLC were downloaded from The Cancer Genome Atlas (TCGA) database to analyze the relation between the expressions of lncRNAs and pN2 nodal metastases. Patients with incomplete clinical information were excluded. This study conforms to the publication guidelines provided by TCGA (https://cancergenome.nih.gov/).
Clinical characteristics and prognosis of T1 NSCLC patients
Pathological T1N0-2M0 NSCLC [LUAD and lung squamous cell carcinoma (LUSC)] patients who underwent surgical resection (lobectomy or pneumonectomy) were screened and allocated to one of three groups according to whether they received MLND and their pN stage: no-MLND, MLND + pN0-1, and MLND + pN2 groups. Firstly, we analyzed the difference of clinical characteristics and postoperative prognoses among groups. The prognosis of patients in the three groups was also analyzed in LUAD patients’ subset. These analyses were performed using the “survival” and “survminer” packages in the R program (https://r-pkgs.org/).
Dataset processing and screening of differentially expressed lncRNAs
After analyzing the transcriptome data from TCGA database, pT1N0-2M0 stage patients were screened and divided into two groups according to their N stage: pN0–N1 and pN2. Differentially expressed lncRNAs were screened by expression difference analysis between pN0-N1 and pN2 patients using limma analysis (Sangerbox; www.sangerbox.com), and the lncRNAs were identified using Ensembl IDs (http://asia.ensembl.org). The expression of N2-related lncRNAs was visualized by heatmap using TBtools software (a toolkit for biologists integrating various biological data-handling tools, https://www.tbtools.com/) (15).
Clinical analysis of lncRNA expression and the diagnostic model construction
In order to explore the correlation between the screened lncRNAs and clinical characteristics, the expression level of the selected lncRNAs in tumor tissues and normal tissues and their correlation with sex, age, and smoking history were analyzed using IBM SPSS Statistics and GraphPad Prism (GraphPad Software). Then, to verify the diagnostic efficacy to detect pN2 metastases of the five selected lncRNAs, we separately analyzed their expressions in pT1 LUAD patients using receiver operating characteristic (ROC) curves. Finally, a combined diagnostic model was constructed using logistic regression and ROC curve analyses.
Correlation analysis of screened lncRNAs and prognosis
The correlations between the screened lncRNAs and overall survival in LUAD were analyzed using the online database “starBase” (https://starbase.sysu.edu.cn/index.php) (16).
Statistical analysis
Statistical analyses for SEER were performed using the R statistical analysis package. Clinicopathological characteristics were analyzed by Chi-Square. Survival analysis was performed using the “survival” and “survminer” packages in the R program (https://r-pkgs.org/). The ROC curves were plotted to evaluate diagnostic efficiency of lncRNAs, and a combined diagnostic model was constructed using logistic regression and ROC curve analyses using SPSS software. An AUC greater than 70% indicates an acceptable model.
Results
Clinical characteristics
From the SEER database, a total of 19,471 patients met inclusion criteria, including 14,146 LUAD cases and 5,325 LUSC cases. Of these, 16,772 patients received MLND (MLND group), and 2,699 cases did not receive MLND (no-MLND group). Additionally, the patients in MLND group included 1,023 cases with pN2 metastasis (6.10%) and 15,749 cases without pN2 metastasis (93.90%) (Table 1). Among the LUAD patients, 12,286 cases received MLND [829 cases with pN2 (6.75%) and 11,457 cases without pN2 (93.25%)], and 1860 cases were classified into no-MLND group.
Table 1
| Characteristics | No-MLND (N=2,699) | MLND | P value | |
|---|---|---|---|---|
| pN0-1 (N=15,749) | pN2 (N=1,023) | |||
| Sex, n (%) | 0.034 | |||
| Male | 1,225 (45.39) | 7,007 (44.49) | 490 (47.90) | |
| Female | 1,474 (54.61) | 8,742 (55.51) | 533 (52.10) | |
| Age, years, mean ± SD | 70.25±9.57 | 68.07±9.05 | 65.78±9.41 | <0.001 |
| Histologic type, n (%) | <0.001 | |||
| Adenocarcinoma | 1,860 (68.91) | 11,457 (72.75) | 829 (81.04) | |
| Squamous cell carcinoma | 839 (31.09) | 4,292 (27.25) | 194 (18.96) | |
| Site, n (%) | 0.129 | |||
| Main bronchus | 3 (0.11) | 10 (0.06) | 1 (0.10) | |
| Upper lobe | 1,637 (60.65) | 9,702 (61.60) | 662 (64.71) | |
| Middle lobe | 141 (5.22) | 821 (5.21) | 61 (5.96) | |
| Lower lobe | 913 (33.83) | 5,166 (32.80) | 296 (28.93) | |
| Overlapping lesion of the lung | 5 (0.19) | 50 (0.32) | 3 (0.29) | |
| Laterality, n (%) | 0.268 | |||
| Left | 1,153 (42.72) | 6,466 (41.06) | 438 (42.82) | |
| Right | 1,546 (57.28) | 9,283 (58.94) | 585 (57.18) | |
| Tumor size, n (%) | <0.001 | |||
| T1a | 663 (24.56) | 1,893 (12.02) | 76 (7.43) | |
| T1b | 1,424 (52.76) | 8,082 (51.32) | 438 (42.82) | |
| T1c | 612 (22.68) | 5,774 (36.66) | 509 (49.75) | |
| Ethnicity, n (%) | 0.078 | |||
| American Indian/Alaska Native | 15 (0.56) | 68 (0.43) | 7 (0.68) | |
| Asian or Pacific Islander | 139 (5.15) | 981 (6.23) | 56 (5.47) | |
| Black | 208 (7.71) | 1,379 (8.76) | 110 (10.75) | |
| White | 2,337 (86.59) | 13,321 (84.58) | 850 (83.09) | |
MLND, mediastinal lymph node dissection.
Differential prognosis of the no-MLND, MLND + pN0-1, and MLND + pN2 groups in pT1 NSCLC
For pT1 NSCLC patients, the MLND patients had a better prognosis than those of the no-MLND patients. Among the MLND patients, those with pN2 metastasis displayed the poorest prognosis. Furthermore, the prognosis of the MLND and no-MLND patients was compared separately for LUAD. These results indicated that the prognosis of the no-MLND group lay between that of the pN0-N1 and pN2 groups (Figure 1).

Correlation of pN2 metastasis with basic clinical features in pT1 NSCLC
The correlational analysis of pN2 metastasis with basic clinical features showed that pN2 metastasis was correlated with age, histologic type, and tumor size, but not with sex, site, laterality, or race. LUAD and patients with larger tumors were more prone to N2 lymph node metastasis (Table 1).
Clinical correlation of screened lncRNAs
As the heatmap shows (Figure 2A), five novel lncRNAs were screened in the pN0–1 and pN2 groups of pT1N0-2M0 LUAD patients: ENSG00000233093 (LINC00892), ENSG00000272525 (AC099522.2), ENSG00000257815 (LINC01481), ENSG00000245556 (SCAMP1-AS1), and ENSG00000277283 (AC004812.2). Then, the correlation of these five lncRNA’s expression with sex, age, and smoking history was analyzed. The results showed that LINC00892 expression levels correlated with age, and AC004812.2 with sex, although patients ≤65 years old and female patients had lower expressions, respectively (Table 2). The expression of LINC00892 was lower (P=0.0009), and AC099522.2 was higher (P=0.0043) in tumor tissues than in normal tissues, whereas LINC01481, SCAMP1-AS1, and AC004812.2 showed no significant differences (P>0.05) (Figure 2B-2F, Table S1). Additionally, in pT1 LUAD, the expression of the five lncRNAs was significantly lower in pN2 patients than in pN0-1 patients (Figure 2G-2K).
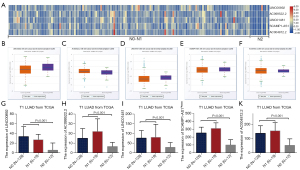
Table 2
| Characteristics | LINC00892 | AC099522.2 | LINC01481 | SCAMP1-AS1 | AC004812.2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median (IQR) | P | Median (IQR) | P | Median (IQR) | P | Median (IQR) | P | Median (IQR) | P | |||||
| Sex | 0.428 | 0.371 | 0.056 | 0.057 | 0.043 | |||||||||
| Male | 28.0 (14.0–44.0) | 16.0 (8.0–26.0) | 78.0 (58.0–118.0) | 285.0 (198.0–359.0) | 160.0 (100.0–198.0) | |||||||||
| Female | 32.0 (15.0–55.0) | 12.5 (8.0–24.8) | 61.5 (45.0–115.5) | 233.5 (159.3–324.8) | 124.0 (80.3–171.8) | |||||||||
| Age, years | 0.017 | 0.256 | 0.329 | 0.714 | 0.080 | |||||||||
| ≤65 | 25.0 (10.5–46.0) | 15.0 (8.0–27.5) | 78.0 (48.5–131.0) | 254.0 (172.5–324.5) | 150.0 (94.0–221.5) | |||||||||
| >65 | 36.0 (20.0–56.3) | 12.5 (8.0–21.5) | 71.0 (45.0–111.3) | 246.0 (176.8–344.3) | 126.0 (84.8–165.0) | |||||||||
| Smokers | 0.805 | 0.277 | 0.238 | 0.224 | 0.082 | |||||||||
| Yes | 28.0 (14.8–53.3) | 15.0 (8.0–26.0) | 75.0 (49.0–127.0) | 264.5 (180.0–342.5) | 140.5 (89.0–198.8) | |||||||||
| No | 33.0 (13.5–54.0) | 12.0 (7.0–21.5) | 64.0 (45.0–113.0) | 221.0 (165.5–315.5) | 115.0 (80.0–164.5) | |||||||||
lncRNAs, long non-coding RNAs; LUAD, lung adenocarcinoma; IQR, interquartile range.
Diagnostic value of the five lncRNAs in pT1 LUAD patients
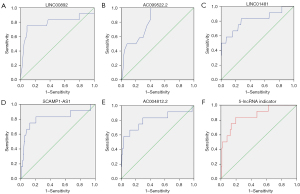
The ROC curve analysis revealed that the areas under curve (AUCs) for the five lncRNAs were as follows: LINC00892, 0.788; AC099522.2, 0.811; LINC01481, 0.812; SCAMP1-AS1, 0.815; AC004812.2, 0.801. The five lncRNAs were then combined to establish a diagnostic model, demonstrating an improved diagnostic efficacy with an AUC of 0.857. The optimum cutoff value showed a sensitivity of 83.3% with a specificity of approximately 80.3% (Figure 3A-3F).
Assessment of the prognostic value of the five lncRNAs in LUAD
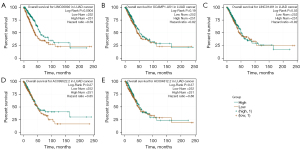
The prognostic values of the five screened lncRNAs were analyzed in LUAD using starBase. The results showed that LINC00892 was correlated with overall survival (OS), with a higher expression level suggesting a better prognosis. However, the remaining four lncRNAs showed no correlation with OS (Figure 4A-4E).
Discussion
MLND has been an important part of curative surgery for lung cancer since the 1990s (17,18). MLND benefits cancer patients but may also increases the risk of postoperative complications, not to mention operative time and cost. There are also suggestions that after immunotherapy, preserving negative lymph nodes may be preferable for early-stage NSCLC patients (19-21). Importantly, consistent with the previous study, we found the prognosis of patients who received MLND was better than that of patients who did not receive MLND, the prognosis of patients with pN2 disease was poorer than that of patients with pN0-N1disease, and the prognosis of patients with no MLND lay between the pN0-N1 and pN2 stage patients in pT1 NSCLC (3). These prognostic results indicate that positive N2 results may have been missed in patients who had not received MLND, leading to their poor prognosis and reinforcing the necessity for MLND in T1 NSCLC. However, a “one-size-fits-all” approach to lymph node dissection may lead to overtreatment, as only 6.10% of pT1 NSCLC patients and 6.75% of pT1 LUAD patients had N2 metastasis. MLND can be used as a targeted treatment if N2 metastasis status can be predicted before surgery.
Further consistent with previous investigators’ findings, our results indicated that the incidence of N2 metastasis correlated with tumor size and histologic type, with LUAD and larger tumor sizes more likely to metastasize (22). However, these basic clinical features are not sufficient for diagnosis. At present, CT, PET/CT, mediastinoscopy, and EBUS-TBNA are the main examination methods for preoperative prediction of N2 status, but they all have limitations. Imaging may show false results due to inflammation and micrometastasis, and mediastinoscopy and EBUS-TBNA are invasive and can result in incomplete sampling. Intraoperative assessment of nodal staging on visual inspection alone is clearly flawed and inadequate (23). Diagnostic biomarkers predicting N2 status have the potential to be of great utility for T1 NSCLC.
Differentially expressed and tissue-specific lncRNAs may be accurate diagnostic markers. In this study, we explored the potential of lncRNAs to be predictive markers of N2 status. After screening, five lncRNAs were identified as possible biomarkers: LINC00892, AC099522.2, LINC01481, SCAMP1-AS1, and AC004812.2. All five lncRNAs had lower expressions in pN2 than pN0–N1 patients and no expression difference between tumor and normal tissues. By analyzing their correlation with basic clinical features, we demonstrated that the five lncRNA expression levels were almost not significantly associated with sex, age, or smoking history.
Additionally, the ROC curve analysis results showed high AUCs (0.788–0.815) for individual lncRNAs. The diagnostic model combining all five lncRNAs showed even better results, with an AUC of 0.857. However, our survival analysis showed that the expression of the five lncRNAs was not closely correlated with OS.
Previous studies have indicated that the sensitivity of CT for detecting N2 metastases is 57%, with a specificity of approximately 82%, and PET/CT demonstrates a high specificity (90%) but a low sensitivity (68%) (22,24). The diagnostic efficiency of CT and PET/CT may also be decreased in early-stage NSCLC. In our study, a combined five-lncRNA diagnostic model showed high specificity and sensitivity, although this will require further verification. But due to the lack of imaging information in the SEER and TCGA databases, we could not establish a combined diagnostic model. However, we suggest that combining lncRNAs and imaging information would further improve diagnostic efficacy and be a powerful reference for surgical treatment. Finally, this study also had some limitations, pathological stage data rather than clinical stage was extracted from SEER database to analyze, but the latter is the main basis for whether MLND is performed. Besides, obtaining tumor tissue samples and rapid analyses of lncRNAs before surgery are the two major challenges at present. And analyses of lncRNAs using liquid biopsy or using small specimens by TBLB (transbronchial lung biopsy)/TNLB (transthoracic needle lung biopsy) may be better for prediction, but still need to be explored.
Conclusions
MLND may be oncologically necessary for selected T1 NSCLC patients based on the metastasis incidence and prognosis. A diagnostic model combining LINC00892, AC099522.2, LINC01481, SCAMP1-AS1, and AC004812.2 expression levels has the potential to be a diagnostic biomarker for N2 metastasis in pT1 LUAD. This study suggests that MLND might be omitted in patients with lower expression level of this diagnostic model. To establish the diagnostic value of this model, further research of the correlation between lncRNA expression and pN2 metastases and the further development of technology detecting lncRNA expression are needed.
Acknowledgments
The authors appreciate the academic support from the AME Thoracic Surgery Collaborative Group.
Funding: This study was funded by The National Natural Science Foundation of China (No. 61801477).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-207/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-207/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All data in this study were downloaded from the public databases, SEER and TCGA.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Gallina FT, Melis E, Forcella D, et al. Minimally invasive hilum-mediastinal lymph nodes dissection for non-small cell lung cancer surgery: What is the future direction? Eur J Surg Oncol 2021;47:1808-9. [Crossref] [PubMed]
- Ray MA, Smeltzer MP, Faris NR, et al. Survival After Mediastinal Node Dissection, Systematic Sampling, or Neither for Early Stage NSCLC. J Thorac Oncol 2020;15:1670-81. [Crossref] [PubMed]
- De Leyn P, Dooms C, Kuzdzal J, et al. Revised ESTS guidelines for preoperative mediastinal lymph node staging for non-small-cell lung cancer. Eur J Cardiothorac Surg 2014;45:787-98. [Crossref] [PubMed]
- Statello L, Guo CJ, Chen LL, et al. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 2021;22:96-118. [Crossref] [PubMed]
- Loewen G, Jayawickramarajah J, Zhuo Y, et al. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol 2014;7:90. [Crossref] [PubMed]
- Liu W, Yin NC, Liu H, et al. Cav-1 promote lung cancer cell proliferation and invasion through lncRNA HOTAIR. Gene 2018;641:335-40. [Crossref] [PubMed]
- Zheng F, Li J, Ma C, et al. Novel regulation of miR-34a-5p and HOTAIR by the combination of berberine and gefitinib leading to inhibition of EMT in human lung cancer. J Cell Mol Med 2020;24:5578-92. [Crossref] [PubMed]
- Pan Y, Liu L, Cheng Y, et al. Amplified LncRNA PVT1 promotes lung cancer proliferation and metastasis by facilitating VEGFC expression. Biochem Cell Biol 2020;98:676-82. [Crossref] [PubMed]
- Xi Y, Shen W, Jin C, et al. PVT1 Promotes the Proliferation and Migration of Non-Small Cell Lung Cancer via Regulating miR-148/RAB34 Signal Axis. Onco Targets Ther 2020;13:1819-32. [Crossref] [PubMed]
- Hua Q, Jin M, Mi B, et al. LINC01123, a c-Myc-activated long non-coding RNA, promotes proliferation and aerobic glycolysis of non-small cell lung cancer through miR-199a-5p/c-Myc axis. J Hematol Oncol 2019;12:91. [Crossref] [PubMed]
- Li Z, Hou P, Fan D, et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ 2017;24:59-71. [Crossref] [PubMed]
- Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res 2017;77:3965-81. [Crossref] [PubMed]
- Doll KM, Rademaker A, Sosa JA. Practical Guide to Surgical Data Sets: Surveillance, Epidemiology, and End Results (SEER) Database. JAMA Surg 2018;153:588-9. [Crossref] [PubMed]
- Chen C, Chen H, Zhang Y, et al. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant 2020;13:1194-202. [Crossref] [PubMed]
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42:D92-7. [Crossref] [PubMed]
- Ochsner A, Debakey M. Primary pulmonary malignancy: treatment by total pneumonectomy; analysis of 79 collected cases and presentation of 7 personal cases. Ochsner J 1999;1:109-25. [PubMed]
- Naruke T, Goya T, Tsuchiya R, et al. The importance of surgery to non-small cell carcinoma of lung with mediastinal lymph node metastasis. Ann Thorac Surg 1988;46:603-10. [Crossref] [PubMed]
- van Pul KM, Fransen MF, van de Ven R, et al. Immunotherapy Goes Local: The Central Role of Lymph Nodes in Driving Tumor Infiltration and Efficacy. Front Immunol 2021;12:643291. [Crossref] [PubMed]
- Fransen MF, Schoonderwoerd M, Knopf P, et al. Tumor-draining lymph nodes are pivotal in PD-1/PD-L1 checkpoint therapy. JCI Insight 2018;3:124507. [Crossref] [PubMed]
- Dammeijer F, van Gulijk M, Mulder EE, et al. The PD-1/PD-L1-Checkpoint Restrains T cell Immunity in Tumor-Draining Lymph Nodes. Cancer Cell 2020;38:685-700.e8. [Crossref] [PubMed]
- Farjah F, Lou F, Sima C, et al. A prediction model for pathologic N2 disease in lung cancer patients with a negative mediastinum by positron emission tomography. J Thorac Oncol 2013;8:1170-80. [Crossref] [PubMed]
- Gaer JA, Goldstraw P. Intraoperative assessment of nodal staging at thoracotomy for carcinoma of the bronchus. Eur J Cardiothorac Surg 1990;4:207-10. [Crossref] [PubMed]
- Xia Y, Zhang B, Zhang H, et al. Evaluation of lymph node metastasis in lung cancer: who is the chief justice? J Thorac Dis 2015;7:S231-7. [PubMed]


