
Peripheral blood leukocyte mitochondrial DNA content and risk of lung cancer
Introduction
Mitochondria are subcellular organelles that produce the majority of cellular ATP through oxidative phosphorylation. Mitochondrial DNA (mtDNA) is a circular, double-stranded molecule comprising 16,569 base pairs that encode 37 genes, including 13 peptides involved in oxidative phosphorylation, two ribosomal RNAs (12S and 16S), and 22 tRNAs (1,2). The mitochondrial genome has several characteristics that differ from the nuclear genome: it is maternally inherited, it lacks introns and protective histones, and has limited DNA repair capacity. The increased susceptibility of mtDNA to damage results in a mutation frequency much higher than that of nuclear DNA (3,4).
Mutations of mtDNA are suspected to be due to the close spatial proximity of the mtDNA genome to the oxidative phosphorylation system, which is located on the inner mitochondrial membrane. mtDNA is thus susceptible to damage through leakage of reactive oxygen species (ROS) during oxidative phosphorylation (5). Some have hypothesized that, in order to maintain the cell’s viability, mitochondria compensate for mutations by increasing mtDNA content (6,7). It has also been theorized that there is a level of ROS exposure beyond which this compensatory mechanism is unable to function, leading to decreases in mtDNA content at very high levels of carcinogen exposure (8). This would suggest that alterations in mtDNA content may be a marker for heightened exposure to various exogenous or endogenous carcinogens (e.g., tobacco smoke) resulting in an increased risk of cancer development.
Changes in mtDNA content have been reported in a wide variety of different cancers, with both increases and decreases described in tumor tissue, body fluids, or peripheral white blood cells (7,9-14). Given the conflicting findings from these previous studies, we evaluated mtDNA content in peripheral blood leukocytes in patients enrolled in a lung cancer case-control study to further evaluate the effect of mtDNA content on the risk of developing lung cancer, and to examine the effect across various levels of tobacco use and different races. We further assessed whether mtDNA content is a prognostic factor in patients with lung cancer who have undergone curative intent resection. We present the following article in accordance with the REMARK reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-21-979/rc).
Methods
Study population
All study subjects were selected from within a case-control study of genetic risk factors for lung cancer at the University of Pennsylvania Health System (Penn). Power calculations were not performed specifically for this study, but sample size was deemed to be adequate in comparison to existing studies of mtDNA content and cancer risk. All study subjects completed a detailed questionnaire and provided a peripheral blood sample prior to initiation of therapy. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All subjects provided informed consent for participation in this study under a protocol approved by the University of Pennsylvania Institutional Review Board (IRB Approval Number: 806390).
Cases were patients with incident lung cancer recruited through medical oncology, thoracic surgery, and pulmonary clinics between 2007 and 2012. Case status was confirmed by reviewing medical records using a standardized form and included only patients with histologically confirmed non-small cell lung cancer (NSCLC) or small cell lung cancer (SCLC). Patients who had a prior diagnosis of cancer at any site other than non-melanoma skin cancer were excluded.
Control subjects were ascertained concurrently with lung cancer cases. Controls were recruited from general medicine and pulmonary clinics. Control subjects were excluded if they had a history of any pulmonary malignancy. A standardized questionnaire was used to collect demographic information, tobacco use history, prior medical history, and other risk factors from cases and controls.
Clinical outcome data were collected for the subset of patients with stage I–II NSCLC that underwent curative intent surgery. In order to better standardize the patient sample for survival analysis, patients undergoing neoadjuvant chemotherapy or radiation were excluded from the study, as were patients who died within 30 days of surgery or had positive resection margins. Outcome data, including information on time to recurrence, recurrence site, and date of death, were obtained via chart review, direct patient contact, local cancer registry data, and from the Social Security Death Index (SSDI).
Measurement of mtDNA content
Whole blood samples were collected from study participants at the time of enrollment. Genomic DNA was extracted from whole blood by use of QIAamp DNA Mini kits (Qiagen). For the determination of mtDNA content relative to nuclear DNA, a quantitative real-time polymerase chain reaction (PCR)-based method was used as previously described, with some modifications (15). One primer pair was used for the amplification of a mitochondrial gene, MT-ND1: forward primer (ND1-F), 5'-CCCTAAAACCCGCCACATCT-3'; reverse primer (ND1-R), 5'-GAGCGATGGTGAGAGCTAAGGT-3'. Another primer pair was used for the amplification of human globulin (HGB), a single-copy nuclear gene: forward primer (HGB-1), 5'-GTGCACCTGACTCCTGAGGAGA-3'; reverse primer (HGB-2), 5'-CCTTGATACCAACCTGCCCAG-3'. The ratio of ND1 copy number to HGB copy number, which is proportional to the mtDNA copy number in each cell, was determined for each sample from standard curves. The ratio for each sample was then normalized to a calibrator DNA sequence in order to standardize between different plates. The PCR reaction was performed in a total volume of 20 µL containing 10 µL Fast 2× SYBR Green Mastermix (Applied Biosystems), 2.5 µL ND1-F (or HGB-1) primer, 2.5 µL ND1-R (or HGB-2) primer, and 5 µL of genomic DNA. The thermal cycling conditions for the mtDNA (MT-ND1 gene) amplification were 95 ℃ for 10 minutes, followed by 40 cycles of 95 ℃ for 15 seconds, and 60 ℃ for 1 minute. The cycling conditions for HGB amplification were 95 ℃ for 10 minutes, followed by 40 cycles of 95 ℃ for 15 seconds, and 56 ℃ for 1 minute. All samples were assayed in duplicate on a 384-well plate with an Applied Biosystems 7900 Sequence Detection System. The PCR runs for ND1 and HGB were always performed on separate plates, but samples from a specific subject were assayed in the same well positions to avoid possible position effect.
A standard curve of a diluted reference DNA was included in each PCR batch to confirm linear assay response and determine copy number, and consisted of a reference DNA sample that was serially diluted to produce a six-point standard curve between 0.3125 and 10 ng of DNA. The R2 for each standard curve was 0.98 or greater. Any sample runs with standard deviations for the cycle of threshold value >0.25 were repeated, and all repeat assays passed this quality control cutoff.
Statistical analysis
The distribution of continuous variables (mtDNA content, age, pack-years of smoking, cigarettes per day, years smoked) was assessed by histograms, calculation of skewness and kurtosis, and Q-Q plots. The distribution of mtDNA among all study participants was unimodal but with moderate deviation from a normal distribution. Hence, mtDNA was natural log-transformed to meet underlying modeling assumptions where mtDNA was modeled as a continuous variable.
We compared distributions of selected case and control variables using the Wilcoxon rank-sum test, the Student’s t-test for continuous variables, and the Pearson chi-squared test for categorical variables. Among case and control subjects separately, we compared mtDNA content (log-transformed) by selected characteristics. We assessed the correlation between mtDNA content and other continuous predictor variables (e.g., age, tobacco pack years) using Spearman correlation coefficients. The relationship between mtDNA content (log-transformed) and tobacco use (categorized as never, previous, and current) was assessed with one-way analysis of variance (ANOVA) followed by Bonferonni corrected pair-wise post-hoc comparisons.
To assess the association between mtDNA content and risk of lung cancer, we used multivariable logistic regression models to determine odds ratios (ORs) by quartile of mtDNA, and with mtDNA considered as a continuous variable. The models were adjusted for age at diagnosis (continuous variable), gender, tobacco use (never, previous, current), and race (White, Black, other). In alternate models, we adjusted for tobacco use by considering cigarettes per day, years smoked, and tobacco pack years. Pre-specified interactions were evaluated between mtDNA content and tobacco use, race, or gender. Stratified models were constructed to specifically examine effects of mtDNA on lung cancer risk by race. The associations between mtDNA content and recurrence-free survival (RFS) or overall survival (OS) were estimated using the method of Kaplan and Meier and assessed using the log-rank test. Cox regression models were used to adjust for potential confounders, with mtDNA content fitted as a two-level variable defined by the median (high vs. low). All analyses were conducted in Stata version 12.
Results
The characteristics of the case and control subjects differed significantly with respect to age, tobacco use, gender, and racial distribution (Table 1). As expected, cases smoked more cigarettes per day, more total years, and had a higher pack year total when compared to controls. The median mtDNA content level for cases was 1.26 [interquartile range (IQR), 0.98–1.70] and 1.79 (IQR, 1.34–2.10) for controls (P<0.001).
Table 1
| Characteristics | Cases (n=465) | Controls (n=378) | P value |
|---|---|---|---|
| Age (years), median [IQR] | 66 [60–73] | 58 [52–64] | <0.0012 |
| Gender, n (%) | 0.21 | ||
| Male | 259 (55.7) | 194 (51.3) | |
| Female | 206 (44.3) | 184 (48.7) | |
| Race, n (%) | <0.0011 | ||
| White | 378 (81.3) | 241 (63.8) | |
| Black | 78 (16.8) | 120 (31.8) | |
| Other | 9 (1.9) | 17 (4.5) | |
| Tobacco use, n (%) | <0.0011 | ||
| Never | 45 (9.7) | 92 (24.3) | |
| Previous | 337 (72.5) | 160 (42.3) | |
| Current | 83 (17.9) | 126 (33.3) | |
| Cigarettes per day**, median [IQR] | 20 [20–30] | 20 [10–30] | <0.0012 |
| Years smoked, median [IQR] | 38 [30–45] | 30 [18–40] | <0.0012 |
| Pack years*, median [IQR] | 41 [25–63] | 27 [13–44] | <0.0012 |
| mtDNA content, median [IQR] | 1.26 [0.98–1.70] | 1.79 [1.34–2.10] | <0.0012 |
| mtDNA content, mean (±SD) | 1.38 (±0.59) | 1.83 (±0.74) | <0.0013 |
Specific cutpoints cigarettes per day, years smoked, and pack years established based on distribution among control subjects. *, data available for 669 of 706 ever smokers; **, data available for 684 of 706 ever smokers; 1, chi-square; 2, rank-sum; 3, Student’s t-test (using natural log transformed data). IQR, interquartile range; mtDNA, mitochondrial DNA; SD, standard deviation.
Effect of tobacco use and race on mtDNA content
Table 2 describes the mtDNA content based on various characteristics of the case and control subjects. mtDNA content did not vary by tobacco history in case subjects but among control participants, mtDNA content among lifelong non-smokers was higher compared to current smokers, a trend which closely approached statistical significance (P=0.05). Among current or previous smokers, mtDNA content had a weak negative correlation with overall tobacco pack years (spearman rho −0.13; P<0.001) and total years smoked (spearman rho −0.09; P<0.001). Although mtDNA content did not vary by race among cases, there was significantly lower mtDNA content among White control subjects compared to Black control subjects (P<0.001).
Table 2
| Variables | Cases (n=465) | Controls (n=378) | |||
|---|---|---|---|---|---|
| No. of subjects | mtDNA content, mean (±SD) | No. of subjects | mtDNA content, mean (±SD) | ||
| Age (years) | |||||
| <58 | 87 | 1.42 (±0.65) | 180 | 1.90 (±0.80) | |
| ≥58 | 378 | 1.38 (±0.57) | 198 | 1.76 (±0.68) | |
| P value1 | 0.64 | 0.10 | |||
| Gender | |||||
| Male | 259 | 1.39 (±0.58) | 194 | 1.80 (±0.70) | |
| Female | 206 | 1.37 (±0.60) | 184 | 1.85 (±0.79) | |
| P value2 | 0.76 | 0.60 | |||
| Race | |||||
| White | 378 | 1.37 (±0.59) | 241 | 1.67 (±0.67) | |
| Black | 78 | 1.43 (±0.56) | 120 | 2.12 (±0.78) | |
| Other | 9 | 1.61 (±0.78) | 17 | 1.87 (±0.83) | |
| P value1 | 0.32 | <0.001* | |||
| Tobacco use | |||||
| Never | 45 | 1.39 (±0.45) | 92 | 1.92 (±0.65) | |
| Previous | 337 | 1.38 (±0.59) | 160 | 1.83 (±0.69) | |
| Current | 83 | 1.39 (±0.64) | 126 | 1.75 (±0.86) | |
| P value1 | 0.89 | 0.05 | |||
| Pack years | |||||
| <27 pack years | 100 | 1.41 (±0.58) | 136 | 1.86 (±0.82) | |
| ≥27 pack years | 365 | 1.38 (±0.59) | 242 | 1.80 (±0.70) | |
| P value2 | 0.57 | 0.61 | |||
| Years smoked | |||||
| <30 years | 91 | 1.39 (±0.67) | 129 | 1.80 (±0.72) | |
| ≥30 years | 374 | 1.38 (±0.57) | 249 | 1.84 (±0.76) | |
| P value2 | 0.92 | 0.66 | |||
| Cigarettes per day | |||||
| ≤20 cigarettes | 239 | 1.40 (±0.58) | 199 | 1.78 (±0.78) | |
| >20 cigarettes | 226 | 1.37 (±0.60) | 179 | 1.88 (±0.70) | |
| P value2 | 0.57 | 0.08 | |||
All analyses performed on log transformed data; raw mean and SD presented. 1, ANOVA; 2, Student’s t-test; *, post-hoc tests resulted in significant difference between Black vs. White (P<0.001). mtDNA, mitochondrial DNA; SD, standard deviation; ANOVA, analysis of variance.
mtDNA content and risk of lung cancer
Lower mtDNA content was significantly associated with lung cancer risk (Table 3). The results were similar in unadjusted models and after controlling for age, race, gender, and tobacco use. Compared to the highest quartile of mtDNA content, there was a significantly increased risk of lung cancer in the second lowest quartile [OR 3.44; 95% confidence interval (CI): 2.06–5.75] and the lowest quartile (OR 6.36; 95% CI: 3.86–10.47).
Table 3
| mtDNA content | Case patients, n | Control subjects, n | Unadjusted OR (95% CI) | Adjusted OR (95% CI) |
|---|---|---|---|---|
| mtDNA content, by quartiles1 | ||||
| Q4 (2.10–5.65) | 37 | 97 | Reference | Reference |
| Q3 (1.67–2.10) | 57 | 92 | 1.49 (0.90–2.47) | 1.57 (0.91–2.73) |
| Q2 (1.24–1.66) | 105 | 96 | 2.87 (1.79–4.60) | 2.85 (1.69–4.78) |
| Q1 (0.32–1.23) | 266 | 93 | 6.89 (4.41–10.77) | 6.44 (3.94–10.54) |
| mtDNA content, by quartiles2 | ||||
| Q4 (2.10–5.65) | 27 | 68 | Reference | Reference |
| Q3 (1.67–2.10) | 50 | 65 | 1.94 (1.09–3.46) | 2.17 (1.15–4.06) |
| Q2 (1.24–1.66) | 86 | 65 | 3.33 (1.92–5.78) | 3.27 (1.81–5.93) |
| Q1 (0.32–1.23) | 232 | 76 | 7.69 (4.59–12.87) | 7.14 (4.07–12.52) |
1, adjusted for age (continuous), race (White, Black, other), gender, and tobacco use (never, previous, current); 2, adjusted for age (continuous), race (White, Black, other), gender, and tobacco pack years (dichotomous). Analysis restricted to current or previous smokers. mtDNA, mitochondrial DNA; OR, odds ratio; CI, confidence interval.
Because tobacco use is the most important risk factor for lung cancer, we performed additional adjustment for this variable. These analyses were performed in a subset of the data where lifetime non-smokers were excluded. Models with additional adjustments for tobacco pack years (Table 3), cigarettes per day (not shown) and years smoked (not shown) did not change the risk estimates appreciably.
Race did not significantly alter the relationship between mtDNA copy number and lung cancer risk (P=0.19). Even though we had a relatively small number of Black subjects, we performed separate analyses by race to further examine any potential differences in lung cancer risk (Table 4). For these analyses, we also dichotomized mtDNA content at the median based on the distribution among control subjects. The risk of lung cancer among Black subjects with low mtDNA copy number was similar when compared to White subjects (OR 4.83 among Blacks vs. OR 3.28 for Whites) after adjustment for age, gender, and tobacco use (Table 4).
Table 4
| mtDNA content, by median1 | Case subjects, n | Control subjects, n | Unadjusted OR (95% CI) | Adjusted OR (95% CI) |
|---|---|---|---|---|
| White and Black patients (n=817) | ||||
| High | 90 | 176 | Reference | Reference |
| Low | 366 | 185 | 3.87 (2.84–5.27) | 3.89 (2.81–5.38) |
| White patients only (n=619) | ||||
| High | 71 | 98 | Reference | Reference |
| Low | 307 | 143 | 2.96 (2.06–4.27) | 3.28 (2.20–4.90) |
| Black patients only (n=198) | ||||
| High | 19 | 78 | Reference | Reference |
| Low | 59 | 42 | 5.77 (3.04–10.92) | 4.83 (2.45–9.51) |
1, adjusted for age (continuous), gender, and tobacco use (never, previous, current). mtDNA, mitochondrial DNA; OR, odds ratio; CI, confidence interval.
OS and RFS
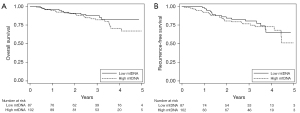
Of the cases included in this study, 189 patients with early-stage NSCLC treated with surgical resection were included to study the effect of mtDNA on RFS and OS. There were a total of 42 cancer recurrences and 34 deaths during the follow-up period. Age, and gender were significantly associated with OS, whereas none of the evaluated variables were associated with RFS. mtDNA copy number was not associated with either RFS (P=0.61) or OS (P=0.32) in univariate analysis (Figure 1). After adjusting for age, gender, and stage, there remained no association between lower mtDNA and either RFS [hazard ratio (HR) 0.89; 95% CI: 0.47–1.66] or OS (HR 0.71; 95% CI: 0.35–1.46).
Discussion
In this case-control study, we show a strong, dose-dependent association between lower levels of mtDNA content in peripheral blood cells and lung cancer. The likelihood of lung cancer was most pronounced in those with the lowest amount of mtDNA content. We also found a lower mtDNA content among current smokers, when compared to either previous or life-long non-smokers, and lower mtDNA content among white control subjects compared to other races. There were no differences in mtDNA content between men and women. Finally, there was no effect of mtDNA on either risk of RFS or OS among patients with early-stage NSCLC treated with surgical resection.
Our results help clarify the existing body of literatures on mtDNA content and risk of lung cancer, which has yielded inconsistent results (Table 5). Our findings are concordant with a recent case-control study nested within two large prospective cohort studies: the Health Professionals Follow-Up Study and the Nurses’ Health Study. In this study, the authors found that among current smokers, the median level of mtDNA content was associated with a higher risk of lung cancer than the high level of mtDNA content (17). These results contrast with a nested case-control study conducted within the Alpha-Tocopherol, Beta-Carotene (ATBC) Cancer Prevention Cohort Study which showed that higher mtDNA copy number was associated with higher risk of lung cancer (10). However, in a pooled analysis where the ATBC study was combined with two additional prospective investigations nested in the Shanghai Women’s Health Study (SWHS) and the Prostate, Lung, Colorectal, and Ovarian (PLCO) cancer screening trial, mtDNA content was not consistently associated with lung cancer risk. Furthermore, an isolated analysis of the PLCO screening trial showed that mtDNA content was inversely associated with lung cancer risk among male smokers (16).
Table 5
| Study | Year | Study type | Number of cases | Number of controls | Method of quantifying mtDNA content | Key findings |
|---|---|---|---|---|---|---|
| Bonner et al. (9) | 2009 | Case-control | 122 | 122 | Sputum mtDNA content determined via qPCR | Higher mtDNA content (>157 copies/cell) was associated with lung cancer risk compared with those with ≤157 copies/cell |
| Hosgood et al. (10) | 2010 | Prospective cohort | 227 | 227 | Whole blood mtDNA content determined via qPCR | Highest quartile of mtDNA content associated with risk of lung cancer compared to lowest quartile |
| Kim et al. (16) | 2014 | Pooled case-control | 880 | 885 | Whole blood mtDNA content determined via qPCR | No consistent evidence of an association across populations by sex or smoking status/intensity |
| Meng et al. (17) | 2016 | Pooled case-control | 463 | 463 | Peripheral blood leukocyte mtDNA content determined via qPCR | Among current smokers, the median level of mtDNA content was associated with a higher risk of lung cancer than the high level of mtDNA content |
| Chen et al. (11) | 2018 | Case-control | 128 | 107 | Plasma mtDNA content determined via qPCR | Lower mtDNA content associated with cases compared to controls |
mtDNA, mitochondrial DNA; qPCR, quantitative polymerase chain reaction.
Cancer sites other than the lung have been studied, including renal cell (15), gastric (18), breast (13), head and neck (14), and non-Hodgkin’s lymphoma (12). These studies also yield inconsistent results for the association between mtDNA content and disease risk. Although there is no clear explanation for the heterogeneity of these results, there are several potential reasons for these inconsistencies. It is possible that some of the differences seen across the various tumor types may be due to differences in study design. Several of these studies, including our own, utilized a case-control design, which raises the possibility that the findings may be due to reverse causation. Some studies suggest that the lower mtDNA content (or mtDNA depletion) may be a consequence of cancer, although these findings are commonly from studies comparing tumor tissues to corresponding normal tissue (19). Previous research in a number of different cancers has provided evidence for two potential mechanisms of mtDNA depletion. The first mechanism involves somatic point mutations in the D-loop region, a 1,124 base-pair stretch of mtDNA that contains essential promotion and replication sequences for mtDNA (20). Mutations in the D-loop region may significantly alter the replicative pattern of mtDNA resulting in reduced tumor mtDNA content (2,7,21,22). mtDNA depletion may also be a consequence of mutations in the p53 pathway. Aberrant expression of p53 may sensitize mtDNA to ROS and alter mtDNA replication (23-27). Although these pathways for mtDNA depletion elucidated in tumor cells have been described, it remains unclear whether solid tumors, such as lung cancer, use these mechanisms to lower mtDNA content in peripheral white blood cells.
In the control arm of our study, we noted significantly lower mtDNA content among current smokers compared to previous or never smokers. This finding is consistent with two recent multi-institutional studies that found a significant decrease in mtDNA content in heavy smokers as compared to former smokers or never smokers (8,17). However, some studies have shown a positive association between tobacco use and mtDNA content (15,28), and others have shown no significant effect (29).
The lack of consistent results in the literature has raised concerns regarding the methodology used to determine mtDNA content (5,30). In most of the analyses performed to date, the methods used to measure the mtDNA content result in data that cannot be directly compared across studies. Studies that have evaluated the variation in mtDNA content measurements across different laboratories suggest some consensus but also significant variability (31,32). Other measurement issues include the potential for duplication of the mitochondrial genome in the nuclear genome, use of inappropriate primers, and template preparation issues (5). It is likely that more robust and reproducible assays are needed to assess mtDNA content. Furthermore, there are a number of issues and uncertainties that prevent mtDNA content from being implemented as a screening biomarker given the current state of knowledge in the field. These issues are summarized in Table 6, with suggested directions for future work in the field.
Table 6
| Barrier to implementing mtDNA testing as a screening biomarker | Putative root causes | Direction for future work |
|---|---|---|
| Inconsistency in associations found between mtDNA content and risk of lung cancer | Variability in study design and mtDNA assays used in different laboratories | Detailed, side-by-side comparison of mtDNA content levels determined by different tests using samples from the same individuals. Ultimately this will help standardize methods and work toward creation of CLIA-certified tests |
| Possibility of reverse causation | Case-control study design | Large prospective cohort studies of mtDNA content and lung cancer risk |
| Uncertainty regarding reproducibility of mtDNA content measurements over time | Case-control study design with only a single measurement of mtDNA content | Longitudinal studies of mtDNA content in the same individuals over time |
| Uncertainty regarding isolated effect of nicotine on leukocyte mtDNA content due to high affinity nicotinic acetylcholine receptors in leukocytes | Difficult to isolate effect of nicotine from other elements of cigarette smoke | Studies on the effect of nicotine from cigarettes compared to nicotine from other sources (nicotine replacement therapy, vaping) on mtDNA content |
| Potential for baseline mtDNA content variation between populations with different environmental exposures, racial/ethnic background, comorbidities, or age | Few existing studies of mtDNA content and lung cancer risk with diverse populations | Further prospective longitudinal studies (particularly in never smokers) in racially and geographically diverse populations |
mtDNA, mitochondrial DNA; CLIA, Clinical Laboratory Improvement Amendments.
Although our study includes a large, racially diverse population of subjects with and without lung cancer, there are several important limitations. The issue of case-control study design was previously discussed, and raises the possibility of reverse causation. In addition, a case-control design does not allow for repeated measures of mtDNA content, as a single measurement may not accurately reflect mtDNA content over the relevant period of disease risk. Second, our study population consists primarily of current or previous smokers, and includes only a small number of never smokers. Because new lung cancers occur only 10–15% of the time in never smokers, this is a difficult population to recruit. Given the potentially important confounding effects of tobacco use on mtDNA variation and content, further studies in never smokers may help determine the effect of other important variables, such as environmental factors, comorbidities, and age on mtDNA content and subsequent disease risk.
The mechanisms involved in the initiation and progression of cancer due to alterations in mtDNA are not completely understood. The risk of lung cancer is greatest among long term current smokers, and an increased risk can persist for decades after smoking cessation. The finding of greater mtDNA depletion among patients with lung cancer and among current smokers supports the possibility that this represents a marker of ongoing “injury” and may identify individuals at highest risk of lung cancer. Further longitudinal, prospective studies are needed to confirm these findings of mtDNA content and lung cancer risk, to elucidate the effect of variation in the mitochondrial genome on mtDNA content, and to study how this variation is modified by smoking and other important risk factors for lung cancer.
Acknowledgments
Funding: This study was supported by NIH-NIEHS grant P30ES013508 and Pennsylvania Department of Health grant PA 4100038714 (to the Center of Excellence in Environmental Toxicology).
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-21-979/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-21-979/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-21-979/coif). GTK reports grant from the National Institutes of Health (Grant F32 CA254210-01) and Daland Fellowship in Clinical Investigation from the American Philosophical Society. TMP, ASW, and AV report grants from the NIH-NIEHS (Grant P30ES013508), and the Pennsylvania Department of Health (Grant PA 4100038714). TMP also reports grant from the NIH-NIEHS (Grant R01ES029294). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the University of Pennsylvania Institutional Review Board (IRB Approval Number: 806390) and all patients gave written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Greaves LC, Reeve AK, Taylor RW, et al. Mitochondrial DNA and disease. J Pathol 2012;226:274-86. [Crossref] [PubMed]
- Yu M. Generation, function and diagnostic value of mitochondrial DNA copy number alterations in human cancers. Life Sci 2011;89:65-71. [Crossref] [PubMed]
- Marcelino LA, Thilly WG. Mitochondrial mutagenesis in human cells and tissues. Mutat Res 1999;434:177-203. [Crossref] [PubMed]
- Kopinski PK, Singh LN, Zhang S, et al. Mitochondrial DNA variation and cancer. Nat Rev Cancer 2021;21:431-45. [Crossref] [PubMed]
- Malik AN, Czajka A. Is mitochondrial DNA content a potential biomarker of mitochondrial dysfunction? Mitochondrion 2013;13:481-92. [Crossref] [PubMed]
- Lee HC, Yin PH, Chi CW, et al. Increase in mitochondrial mass in human fibroblasts under oxidative stress and during replicative cell senescence. J Biomed Sci 2002;9:517-26. [Crossref] [PubMed]
- Lee HC, Li SH, Lin JC, et al. Somatic mutations in the D-loop and decrease in the copy number of mitochondrial DNA in human hepatocellular carcinoma. Mutat Res 2004;547:71-8. [Crossref] [PubMed]
- Wu S, Li X, Meng S, et al. Fruit and vegetable consumption, cigarette smoke, and leukocyte mitochondrial DNA copy number. Am J Clin Nutr 2019;109:424-32. [Crossref] [PubMed]
- Bonner MR, Shen M, Liu CS, et al. Mitochondrial DNA content and lung cancer risk in Xuan Wei, China. Lung Cancer 2009;63:331-4. [Crossref] [PubMed]
- Hosgood HD 3rd, Liu CS, Rothman N, et al. Mitochondrial DNA copy number and lung cancer risk in a prospective cohort study. Carcinogenesis 2010;31:847-9. [Crossref] [PubMed]
- Chen J, Zhang L, Yu X, et al. Clinical application of plasma mitochondrial DNA content in patients with lung cancer. Oncol Lett 2018;16:7074-81. [Crossref] [PubMed]
- Kim C, Bassig BA, Seow WJ, et al. Mitochondrial DNA copy number and chronic lymphocytic leukemia/small lymphocytic lymphoma risk in two prospective studies. Cancer Epidemiol Biomarkers Prev 2015;24:148-53. [Crossref] [PubMed]
- Lemnrau A, Brook MN, Fletcher O, et al. Mitochondrial DNA Copy Number in Peripheral Blood Cells and Risk of Developing Breast Cancer. Cancer Res 2015;75:2844-50. [Crossref] [PubMed]
- Reznik E, Miller ML, Şenbabaoğlu Y, et al. Mitochondrial DNA copy number variation across human cancers. Elife 2016;5:10769. [Crossref] [PubMed]
- Xing J, Chen M, Wood CG, et al. Mitochondrial DNA content: its genetic heritability and association with renal cell carcinoma. J Natl Cancer Inst 2008;100:1104-12. [Crossref] [PubMed]
- Kim C, Bassig BA, Seow WJ, et al. Pooled analysis of mitochondrial DNA copy number and lung cancer risk in three prospective studies. Cancer Epidemiol Biomarkers Prev 2014;23:2977-80. [Crossref] [PubMed]
- Meng S, De Vivo I, Liang L, et al. Pre-diagnostic leukocyte mitochondrial DNA copy number and risk of lung cancer. Oncotarget 2016;7:27307-12. [Crossref] [PubMed]
- Zhu X, Mao Y, Huang T, et al. High mitochondrial DNA copy number was associated with an increased gastric cancer risk in a Chinese population. Mol Carcinog 2017;56:2593-600. [Crossref] [PubMed]
- Schon EA, DiMauro S, Hirano M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat Rev Genet 2012;13:878-90. [Crossref] [PubMed]
- Clayton DA. Transcription and replication of mitochondrial DNA. Hum Reprod 2000;15:11-7. [Crossref] [PubMed]
- Yu M, Zhou Y, Shi Y, et al. Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients. IUBMB Life 2007;59:450-7. [Crossref] [PubMed]
- Yu M, Wan Y, Zou Q. Decreased copy number of mitochondrial DNA in Ewing's sarcoma. Clin Chim Acta 2010;411:679-83. [Crossref] [PubMed]
- Achanta G, Sasaki R, Feng L, et al. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. EMBO J 2005;24:3482-92. [Crossref] [PubMed]
- Chang SC, Lin PC, Yang SH, et al. Mitochondrial D-loop mutation is a common event in colorectal cancers with p53 mutations. Int J Colorectal Dis 2009;24:623-8. [Crossref] [PubMed]
- Kulawiec M, Ayyasamy V, Singh KK. p53 regulates mtDNA copy number and mitocheckpoint pathway. J Carcinog 2009;8:8. [Crossref] [PubMed]
- Lebedeva MA, Eaton JS, Shadel GS. Loss of p53 causes mitochondrial DNA depletion and altered mitochondrial reactive oxygen species homeostasis. Biochim Biophys Acta 2009;1787:328-34. [Crossref] [PubMed]
- Singh KK, Ayyasamy V, Owens KM, et al. Mutations in mitochondrial DNA polymerase-gamma promote breast tumorigenesis. J Hum Genet 2009;54:516-24. [Crossref] [PubMed]
- Masayesva BG, Mambo E, Taylor RJ, et al. Mitochondrial DNA content increase in response to cigarette smoking. Cancer Epidemiol Biomarkers Prev 2006;15:19-24. [Crossref] [PubMed]
- Yang K, Forman MR, Graham BH, et al. Association between pre-diagnostic leukocyte mitochondrial DNA copy number and survival among colorectal cancer patients. Cancer Epidemiol 2020;68:101778. [Crossref] [PubMed]
- Nguyen H, LaFramboise T. Complexities and pitfalls in analyzing and interpreting mitochondrial DNA content in human cancer. J Genet Genomics 2020;47:349-59. [Crossref] [PubMed]
- Côté HC, Gerschenson M, Walker UA, et al. Quality assessment of human mitochondrial DNA quantification: MITONAUTS, an international multicentre survey. Mitochondrion 2011;11:520-7. [Crossref] [PubMed]
- Hammond EL, Sayer D, Nolan D, et al. Assessment of precision and concordance of quantitative mitochondrial DNA assays: a collaborative international quality assurance study. J Clin Virol 2003;27:97-110. [Crossref] [PubMed]


