
Increased serum levels of KiSS1-derived peptides in non-small cell lung cancer patient liquid biopsies and biological relevance
Introduction
Lung cancer is the leading cause of cancer death in the world. Non-small cell lung cancer (NSCLC), the major form, is characterized by late diagnosis and poor prognosis and accounts for more than 85% of all lung cancer (1). In patients lacking actionable mutations, platinum-based therapy is employed in advanced disease, and combined with immunotherapy has shown promising results in clinical trials (2,3). Immunotherapy is now the standard treatment for advanced NSCLC and it is being introduced also at earlier disease stages (4). In spite of this, disease outcome remains unsatisfactory due to drug resistance and metastases (5). Indeed, several factors have been implicated in the drug resistance development, including gene mutations and post-transcriptional modifications. Epigenetically regulated mechanisms may modulate tumor plasticity, allowing to reprogram resistant clones by epigenetic drugs (6), thereby influencing cell response to treatment and aggressive tumor cell features.
A number of molecular alterations [e.g., epidermal growth factor receptor (EGF-R) mutation, anaplastic lymphoma kinase translocation, programmed death ligand 1 level] are currently used as biomarkers to stratify and treat NSCLC patients (5). Biomarker identification most commonly relies on surgical sample collections, whereas minimally invasive liquid biopsies may represent a valid alternative to tissue biopsies, particularly in NSCLC patients. Thus, the analysis of peripherally circulating biomarkers of various origin, offers a new source of materials that may reflect the disease status and thereby may be useful for personalized treatment (7).
KiSS1-derived peptides (i.e., kisspeptins) are the products of the metastasis suppressor gene KiSS1, and may represent promising biomarkers, because they are secreted and endowed with biological activity (8). KiSS1 codes for a 145-aa polypeptide, from which a 54-aa peptide [kisspeptin-54 (KP54)/metastin] proteolitically cleaved into shorter products (kisspeptin-10, KP10; kisspeptin-13, KP13; kisspeptin-14, KP14) is generated (9,10). The KiSS1 gene, originally found to inhibit metastasis of cancer cells through down-regulation of matrix metalloproteinases (11), also plays a key physiological role being involved in the control of puberty and reproductive function (12). The role of KiSS1 signalling in cancer has not been fully elucidated, although it appears to be involved in metastasis control and in response to cisplatin (13,14). It has been demonstrated that KiSS1 blocks metastatic colonization in melanoma, being necessary for metastasis suppression and—when secreted—to maintain dormancy in disseminated cells (8,15). Several studies have linked KiSS1 loss to progression/metastases in various cancer types besides melanoma. We previously found that forced expression of KiSS1 decreased the invasive capability of cisplatin-resistant cells, similarly to the effect of histone deacetylase (HDAC) inhibitors which up-regulated KiSS1 mRNA levels (16). This study highlights a possible link between KiSS1 and epigenetic mechanisms active in drug-resistant cells. Indeed, the relevance of histone acetylation in KiSS1 regulation of drug-resistant cells emerged, together with additional evidence from the literature indicating KiSS1 regulation by DNA methylation or microRNAs (17).
Based on this background, since the role of KiSS1 in NSCLC is not well defined, the aim of the present study was to investigate the possibility of measuring KiSS1 levels in liquid biopsies from NSCLC patients in comparison to healthy donors and to explore the biological significance of KiSS1 in NSCLC experimental models. To this end, we designed a pilot study to analyse KiSS1 levels in liquid biopsies including three biological fluids (serum, plasma and urine) from healthy donors (controls) and NSCLC patients (cases) before and at follow up, thereby exploring potential applications of KiSS1 as a tool to monitor disease features in NSCLC patients. To explore the biological significance of KiSS1 based on our previous work showing epigenetic regulation of KiSS1 upon treatment with HDAC inhibitors (16), we first examined KiSS1 mRNA modulation by the epigenetic agent azacytidine. Then, because KiSS1 is secreted, we examined its levels by ELISA upon pharmacological modulation and, given that KiSS1 has been implicated in regulation of apoptotic response to cisplatin in head and neck cancer (14) we evaluated if KiSS1-derived peptides could increase cisplatin-induced apoptosis. Finally, because KiSS1 is expressed in the hypothalamus (18) and we hypothesized that the development of cancer may increase KiSS1 levels in the organism, KiSS1 production by neurons challenged by tumour cell conditioned medium was assayed. We present the following article in accordance with the MDAR reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-52/rc).
Methods
Ethical statement
The research was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from all subjects upon approval of the study by institutional review board Fondazione IRCCS Istituto Nazionale dei Tumori (protocol number INT 167/16).
Population cohorts
The population consists of a cohort of 60 NSCLC patients (cases) at different stages of disease (I–IV) and 60 healthy donors (controls), matched for sex and smoking habits, consecutively recruited from January 2017 to April 2018 at the Thoracic Surgery Unit and at the Immunohaematology and Transfusion Medicine Service of the Fondazione IRCCS Istituto Nazionale dei Tumori. Fifty-nine patients were matched with 59 healthy donors for sex and smoking habits, as one patient was excluded due to a rare lung sarcoma miming a NSCLC. Patients with a confirmed histological diagnosis of NSCLC were enrolled. For each enrolled subject, demographical and clinical data were recorded. Enrolled patients underwent diagnostic non-invasive surgical procedure or surgical procedure; before performing this, blood and urine sample (at the time of admission, i.e., baseline) were collected. After surgical procedure adjuvant therapy (chemotherapy, radiotherapy, both or none) were performed according to pathological stage and oncologic multidisciplinary evaluation. After 4–6 months from the previous procedure, a radiological and clinical follow-up for all the recruited patients was performed and other blood and urine samples were collected (follow-up samples).
Quantitative analysis of KiSS1 in biological fluids
KiSS1 levels were measured in biological fluids by ELISA kit (Human Metastasis Suppressor KiSS-1 kit, Cusabio, Houston, TX, USA), according to the manufacturer’s instructions for quantitative analysis. Aliquots of urine, plasma and serum samples from patients and healthy subjects were used for the assay in triplicate. Urine levels were normalized with respect to creatinine content. Creatinine plasmatic levels were assessed by the Jaffe method using routine automatic analyzer (Architect, Abbott, Lake Forest, IL, USA). A calibration curve was fitted by plotting the mean plate standard’s absorbance (dependent variable) as a function of the known KiSS1 concentrations of the standard (independent variable). This curve was then used to estimate the unknown starting concentration in the test samples.
Cell lines
The human NSCLC cell lines H460 (NCI-DTP Cat# NCI-H460, RRID: CVCL_0459), the cisplatin-resistant variant H460/Pt and the H1975 cell line (ATCC, NCI-H1975, RRID: CVCL_1511) were grown in RPMI-1640 medium (Lonza, Basel, Switzerland), supplemented with 10% foetal bovine serum (FBS; Gibco, Life Technologies, Carlsbad, CA, USA). The cisplatin-resistant cells were obtained as previously described (16). Cells were routinely checked for mycoplasma contamination (Lonza), used within 20 passages from thawing from a frozen stock. Cell identity was authenticated through microsatellite analysis by the AmpFISTR Identifiler PCR amplification kit (Applied Biosystem, Carlsbad, CA, USA). For in vitro studies, cisplatin (Accord Healthcare Italia, Milan, Italy) and azacytidine (Selleckchem, Aurogene, Rome, Italy) were diluted in saline. The KiSS1-derived peptide KP54 was obtained from Anaspec, DBA Italia (Milan, Italy).
Quantitative real time polymerase chain reaction (qRT-PCR)
Gene expression levels were analyzed by qRT-PCR according to standard methods in untreated cells and in cells exposed to azacytidine for 24 h in 6-well plates, 24 h after seeding. The RNeasy Plus Mini kit (Qiagen, Hilden, Germany) was used to isolate RNA, which was reversed transcribed employing the High Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific, Monza, Italy). The following TaqMan assays were used: Hs.PT.58.2731441 for KiSS1 and Hs02758991_g1 for GAPDH (Thermo Fisher Scientific). Technical triplicate reactions were carried out with a 7900HT Fast Real-Time PCR System (Thermo Fisher Scientific) and data were acquired through the Sequence Detection Systems (SDS) 2.4 software. Reactions were in a 10 µL volume comprising cDNA (2.5 µL), master mix (5 µL, TaqMan Universal Fast PCR Master Mix, Thermo Fisher Scientific) and the specific assay (0.5 µL). By the relative quantification (RQ) manager software (Thermo Fisher Scientific), we determined cDNA relative levels as previously described (19), applying the RQ method, choosing untreated cells as calibrator.
Quantitative analysis of KiSS1 in the culture medium of tumor cells
Cells were seeded at 4×105 cells/dish (19.6 cm2) in complete medium and cultured for 24 h before treatment with azacytidine for 24 h; then cisplatin was added for 48 h. At the end of treatment, culture media were harvested and clarified by centrifugation at 13,000 rpm for 5 min and an aliquot was used for the ELISA (see above). Adherent cells were counted to allow normalization of the KiSS1 peptide level. Three independent experiments were performed and the mean value [± standard deviation (SD)] was calculated.
Apoptosis analyses
Apoptosis was evaluated by measuring the activation of caspase 3/7 using the luminescent Caspase Glo 3/7 assay (Promega, Fitchburg, WI, USA). Cells were seeded in 96 well plates (7,000 cells/well in 100 µL of medium) and 24 h later they were exposed to KP54, cisplatin or to their combination. After 48 h, caspase 3/7 activation was examined following manufacturer’s instructions. Relative luminescence units (RLU) were normalized with respect to the total protein content of each well to correct for the growth inhibitory effect of the treatment. Protein content was assayed by the BCA method.
The annexin V-binding assay (Immunostep, Salamanca, Spain) was also used to evaluate apoptosis in H460, H460/Pt and H1975 cells. Cells were treated for 48 h with KP54, cisplatin or their simultaneous combination. After washing with cold phosphate-buffered saline (PBS), cells were resuspended in a buffer containing 10 mM Hepes-NaOH, pH 7.4, 2.5 mM CaCl2, and 140 mM NaCl (binding buffer). Following incubation of cells (105) with 5 µL of FITC-conjugated annexin V and 10 µL of 2.5 µg/mL propidium iodide at room temperature in the dark for 15 min, annexin V-binding was examined by flow cytometry (BD Accuri, Becton Dickinson, Milan, Italy), acquiring ten thousand events for sample. Results were analyzed using the instrument software (Becton Dickinson).
Human neural stem cell (hNSC) differentiation
hNSCs were derived from 12-week-old healthy forebrains collected after elective routine abortions, following the ethical guidelines of the European Network for Transplantation (NECTAR, https://nectar-eu.com/). The use of human central nervous system brain tissue was approved by the Ethics Committee of the Neurological Institute Foundation ‘Carlo Besta’ and “L. Mangiagalli” Obstetric-Gynecological Clinic of Milan, Italy. Brain tissue mechanical dissociation, primary culturing and cell propagation were carried out as described (20). Cells were seeded in the presence of 20 ng/mL of human recombinant epidermal growth factor (EGF) and 10 ng/mL of basic fibroblast growth factor (FGF) (Peprotech, London, UK) in NeurocultTM—XF Basal Medium for Neural Stem Cells added with Neurocult Proliferation Supplement (Stem Cell Technologies, Vancouver, Canada) and 2 µg/mL heparin (Pharepa, 5,000 U.I.). Cultures were maintained in a humidified incubator at 37 ℃ and 5% CO2. hNSCs neurospheres were dissociated to single cells every 7–10 days with 0.3 PZ U/mL collagenase NB6 (SERVA, Heidelberg, Germany) and replated in the same growth medium at density of 104 cells/cm2. Multipotency ability of hNSCs was determined by in vitro differentiation tests, in which the simultaneous presence of neurons, astrocytes and oligodendrocytes was detected (20,21). Specifically, hNSCs differentiated on coated chamber slide, were fixed for 20 min in 4% paraformaldehyde at 4 ℃, washed and incubated for 30 min at 37 ℃ with PBS plus 0.1% Triton-X containing 10% normal goat serum. Cells were then incubated for 90 min with the appropriate primary antibody diluted in PBS: rabbit Anti-Glial Fibrillary Acid Protein (GFAP, 1:500, Millipore, Burlington, MA, USA, Cat# AB5804), mouse Anti-Tubulin Beta III (1:200, Millipore, Cat# MAB1637; RRID:AB_2210524), or mouse Anti-Galactocerebroside (GalC, 1:200, Millipore, Cat# MAB342; RRID:AB_94857). Cells were washed with PBS and incubated for 1 h at room temperature with the secondary antibody: Goat Anti-Rabbit IgG H&L (Alexa Fluor® 488, 1:1,000, Abcam, Cambridge, UK, ab150077), Goat Anti-Mouse IgG H&L (Alexa Fluor® 594, 1:2,000, Abcam, ab150116) or Goat Anti-Mouse IgG H&L (Alexa Fluor® 488, 1:2,000). After washing, cells were incubated with 4’,6-diamidino-2-phenylindole (DAPI, 0.2 µg/mL) for 10 min at room temperature in the dark and mounted with the FluorSaveTM reagent (Calbiochem, San Diego, CA, USA). Differentiated hNSCs were viewed under an inverted fluorescence microscope (Nikon Eclipse TE300, Nikon, Tokyo, Japan). hNSCs neurospheres were dissociated and plated on Corning Matrigel Basement Membrane Matrix Growth Factor Reduced (Corning, NY, USA) at density of 4×104 cells/cm2 in the same medium used during routine culturing supplemented with 2% FBS w/o growth factors. Cells were incubated for 7–10 days at 37 ℃ and 5% CO2. After 7–10 days cells were treated with conditioned medium obtained by growing NSCLC cells in RPMI-1640 in the absence of serum for 24 h. Supernatants collected after 24 and 48 h were cryopreserved at −80 ℃ and then used for KiSS1 ELISA.
Statistical analysis
The comparison of the distribution of the KiSS1 levels in the considered biological matrices in NSCLC patients at admission (cases) with that of healthy donor (controls) was summarized by descriptive statistics. To assess the association between the KiSS1 levels and the disease status, conditional logistic regression models (22) were performed. The non-parametric Wilcoxon signed rank (WSR) test (22) was applied to compare the distribution of KiSS1 levels in NSCLC patients at baseline and follow-up time (i.e., 4–6 months after admission date) in each biological matrix. The relationship between the tumor stage and the KiSS1 levels in NSCLC patients was investigate by resorting to the Kruskal-Wallis (KW) test (23). The strength of association of KiSS1 levels in each matrix with each of the remaining ones was assessed with the Spearman correlation coefficient (rs) and its 95% confidence interval computed according to the bias-corrected and accelerated (BCa) bootstrap method (95% CIBCa) (24).
For preclinical experiments, the relationship between KiSS1 expression and the different considered factors was investigated by using the non-parametric Wilcoxon (W) or KW tests according to the number of considered groups (22) and corresponding P values were estimated according to exact test or via Monte Carlo approaches. Finally, to jointly evaluate the combination of KP54 with cisplatin on caspase and apoptosis, a two-way analysis of variance (ANOVA) model was implemented by including the main factors and the first order interaction term. Significant interactions were decomposed by stratifying by cisplatin. All statistical analyses were performed with the SAS software (Version 9.4.; SAS Institute Inc., Cary, NC, USA) by adopting a nominal significance level of alpha equal to 0.05.
Results
Clinical study
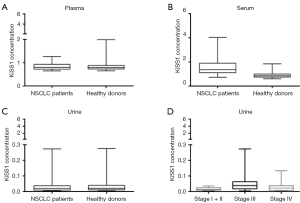
From the 120 enrolled subjects, 74 males and 44 females were assessable, with a median age of 64 years (range, 39–85 years), 58 being currently smokers, 18 never smokers and 42 ex-smokers including patients who had stopped smoking for at least 12 months (25) (Table 1). As regards NSCLC patients, 79.66% of them had an adenocarcinoma, whereas the remaining ones were squamous cell carcinoma and sarcomatoid carcinoma (Table 1). By resorting to conditional regression models, a significant association was found between KiSS1 levels and disease status for the serum matrix (Table 2, Figure 1A-1C). When considering the NSCLC patients only, a significant association between tumor stages (classified as stage I + II, stage III and stage IV) and KiSS1 in the urine matrix was found by KW test. Specifically, as reported in Figure 1D, the distribution of KiSS1 was significantly lower in patients with stage I + II compared with that of stage III (Bonferroni adjusted P value =0.01). Moreover, the distribution of KiSS1 levels in 39 NSCLC patients with both baseline and follow-up samples, significantly decreased in the serum matrix (P value <0.001) at follow-up (Figure 2). Finally, we observed a statistically significant correlation between the plasma and serum matrices by looking at the overall cohort (rs=0.357, 95% CIBCa: 0.184–0.504), within NSCLC patients (rs=0.394, 95% CIBCa: 0.110–0.575) and healthy donors (rs=0.521, 95% CIBCa: 0.273–0.722), even if with a low magnitude. No other significant relationships were observed between the other matrices.
Table 1
| Variables | Healthy donors | NSCLC patients | |||
|---|---|---|---|---|---|
| N | % | N | % | ||
| Sex | |||||
| Female | 22 | 37.29 | 22 | 37.29 | |
| Male | 37 | 62.71 | 37 | 62.71 | |
| Smoking habits | |||||
| Current smoker | 29 | 49.15 | 29 | 49.15 | |
| Ex-smoker | 21 | 35.59 | 21 | 35.59 | |
| Never smoker | 9 | 15.25 | 9 | 15.25 | |
| Tumor stage | |||||
| I + II | – | – | 25 | 42.37 | |
| III | – | – | 21 | 35.59 | |
| IV | – | – | 13 | 22.03 | |
| Histology | |||||
| ADK | – | – | 47 | 79.66 | |
| Squamous cell carcinoma | – | – | 10 | 16.95 | |
| Sarcomatoid carcinoma | – | – | 2 | 3.39 | |
| Age at diagnosis | |||||
| Median (range) | 63 [56–70] | 65 [39–85] | |||
NSCLC, non-small cell lung cancer; ADK, adenocarcinoma.
Table 2
| Matrix | Group | N | Min | 25th centile | Median | 75th centile | Max | IQR1 | OR (95% CI)2 |
|---|---|---|---|---|---|---|---|---|---|
| Plasma | NSCLC patients | 59 | 0.644 | 0.718 | 0.796 | 0.924 | 1.265 | 0.206 | 0.944 (0.607, 1.469) |
| Healthy donors | 59 | 0.647 | 0.728 | 0.786 | 0.873 | 1.983 | 0.145 | – | |
| Serum | NSCLC patients | 59 | 0.725 | 1.109 | 1.352 | 1.895 | 4.023 | 0.786 | 2.939 (1.696, 5.093) |
| Healthy donors | 59 | 0.610 | 0.745 | 0.846 | 0.998 | 1.841 | 0.253 | – | |
| Urine | NSCLC patients | 59 | 0.004 | 0.009 | 0.019 | 0.038 | 0.273 | 0.029 | 0.993 (0.677, 1.456) |
| Healthy donors | 59 | 0.005 | 0.011 | 0.021 | 0.040 | 0.276 | 0.028 | – |
1, IQR: interquartile range, 75th centile–25th centile; 2, odds ratio computed for a specific unit change of the KiSS1 levels equal to one SD computed among controls (SD plasma =0.22; SD serum =0.24; SD urine =0.05) and adjusted by age at baseline. NSCLC, non-small cell lung cancer; CI, confidence interval; SD, standard deviation.

Pharmacological modulation of KiSS1 levels in NSCLC cells
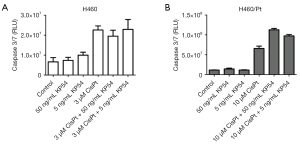
KiSS1 gene expression levels were examined in cisplatin-sensitive H460 and cisplatin-resistant H460/Pt cells after 24 h exposure to the demethylating agent azacytidine by qRT-PCR. In both cell lines, a marked increase of KiSS1 levels was observed (Figure 3A). Since KiSS1 is a secreted peptide (8), we quantitatively assessed the modulation of KiSS1 levels by ELISA in cell culture media derived from H460 and H460/Pt cells (Figure 3B, KW P value =0.002 and 0.0168 for H460 and H460/Pt, respectively). In cells exposed to 1 µM azacytidine for 24 h and then co-incubated with cisplatin (3 µM for H460 and 10 µM for H460/Pt cells) for 48 h, higher concentrations of KiSS1 were released in culture media of both parental and drug-resistant cells as compared to untreated cells. The mRNA levels of KiSS1 in cells exposed to the combination were not increased (data not shown), likely reflecting the occurrence of cell death.

Tumor cell response to KiSS1-derived peptide-cisplatin combinations
Because KiSS1 may modulate cisplatin sensitivity in tumour cells (14), we examined the effect of KiSS1-derived peptide levels on cisplatin-induced apoptosis in NSCLC cell lines by evaluating activation of caspases 3/7 (Figure 4). Using the KiSS1-derived peptide KP54, we observed activation of caspases by cisplatin in the H460/Pt cells but not in cells exposed for 48 h to the peptides per se (P interaction <0.001). Of note, the combination of KP54 with cisplatin markedly increased apoptosis in cisplatin-resistant cells (contrasts P value <0.001).
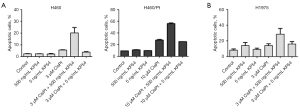
When the effect of the combination was examined using the annexin V-binding assay (Figure 5), higher levels of apoptosis were observed comparing H460 or H460/Pt cells exposed to the combination of KP54 and cisplatin (P interaction <0.01). The combination of KP54 and cisplatin induced a moderate (P interaction =0.14) increase in levels of apoptosis also in the H1975 cell line, characterized by EGF-R L858R/T790M double mutations that confer resistance to targeted therapy. Of note, higher values were observed in the both cisplatin and KP54 factors resulting statistically significant.
KiSS1 production by neuron-enriched cultures
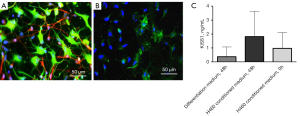
When addressing the reason why cases displayed increased serum KiSS1 levels as compared to healthy donors, we considered that KiSS1 is expressed in the hypothalamus (18) and hypothesized that the development of cancer may increase KiSS1 levels in the organism. To address this point, we differentiated neurons from hNSC (Figure 6A,6B) and we cultured them in the presence of tumor cell-conditioned medium for 48 h. Under these conditions (Figure 6C), we found increased levels of KiSS1 upon exposure of cultures enriched in neurons to H460 cell conditioned medium as compared to differentiation medium (one-sided Wilcoxon P value =0.04). Tumor cells also appeared to produce KiSS1 as shown by the KiSS1 levels found in the conditioned medium prior incubation with neurons (H460 cells, 0 h).
Discussion
KiSS1 levels in liquid biopsies of cancer patients have been poorly investigated until now. In 31 ovarian carcinoma patients (26) plasma kisspeptin concentration, measured using an antibody detecting KP54, KP14 and KP10 (27) was found dependent on the disease stage, with stage I ovarian carcinoma patients displaying increased levels than stage II and IV and healthy controls. KP10 levels were higher in patients with small renal tumours than controls, with different concentrations in subgroups of patients (28). Enhanced levels were also observed for plasma KP54 in patients with gastric cancer and in colorectal cancer at initial stage versus controls (29,30). Overall, these studies are in keeping with our findings of increased KiSS1-derived peptide levels in NSCLC patients versus sex and smoke-habit matched healthy controls. With the test used in the present study, KiSS1 is quantified implying that different KiSS1-derived peptides, i.e., KP54, KP10, KP13 and KP14 are detectable.
Liquid biopsy has quickly moved from research into clinical practice being minimally-invasive and easily repeatable (31). Since a little information is available on KiSS1 in liquid biopsy of cancer patients, we collected samples from three different biological matrices from NSCLC patients, and employed ELISA to measure KiSS1 release. Due to the low molecular weight of KiSS1-derived peptides, they can be detected also in urine, a valuable matrix because samples are collectable in a non-invasive manner.
We found that KiSS1 levels in serum were increased in NSCLC patients as compared to healthy donors. KiSS1 has a well-known role in the regulation of metastatic spread, particularly in its suppression, through maintenance of dormancy of disseminated malignant cells and by interfering with cell migratory and invasive abilities (17). Thus, our result was unexpected and may suggest that when cancer develops, mechanisms counteracting dissemination are concurrently activated. When considering the KiSS1concentration in urine (but not in serum), among NSCLC patients, different levels were observed, depending on disease stage, with higher values for stage III patients. A large heterogeneity in KiSS1 levels was—however—found in stage III patients. Because a fraction of stage III patients received platinum-based neo-adjuvant therapy which may have nephrotoxic effect, higher urine KiSS1 levels are conceivable. A decrease of serum KiSS1-derived peptides was observed at follow-up, suggesting that KiSS1 values were similar to physiological levels in patients likely exhibiting a favorable outcome, given their possibility to have a follow-up sample. A correlation between plasma and serum KiSS1 level was observed in cases and among all samples. Although this observation seems to suggest the utility of all the studied matrices as sources of KiSS1, in the present study we did not find any statistically significant modulation of KiSS1 level in the plasma matrix between NSCLC patients and control subjects. Therefore, according to our pilot study results, serum seems to be the most suitable matrix for determining KiSS1 modulation. Indeed, it seems likely that the antigen-antibody interaction necessary for the ELISA has a better performance in serum.
In cell medium of NSCLC cells sensitive and resistant to cisplatin, we found that KiSS1-derived peptide levels were increased upon exposure to the combination of cisplatin and azacytidine. This observation suggests the opportunity to further investigate KiSS1 modulation in relation to response to therapy, to exploit its role as a predictive biomarker. Of note, in the cellular models, we observed increased cisplatin-induced apoptosis upon co-incubation with KiSS1-derived peptides in cisplatin-resistant H460/Pt cells. Such findings suggest that KiSS1-derived peptide can modulate cisplatin sensitivity in NSCLC cells, in keeping with a report in head and neck cancer (14). In those cells, knockdown of KiSS1 suppressed cisplatin-induced poly-ADP ribose polymerase cleavage and increased glutathione transferase pi protein level and activity (14).
Although it is still unclear why cases displayed increased serum KiSS1 levels as compared to healthy donors, the results obtained when incubating differentiated neurons with NSCLC cell conditioned medium, suggest a possible increase of physiological KiSS1 levels upon cancer development. The effect—despite being significant—may be down-tuned by the presence of additional cell types besides neurons in our experimental model, not necessarily producing KiSS1.
A better understanding of the role of KiSS1 in cancer biology may also benefit from considering KiSS1 protein-protein interactions. In this regard, the best documented one is the interaction with its receptor (17). Besides, bioinformatics approaches such as those available from the STRING interaction network (https://string-db.org) identify Tachykinin-3 as a functional interactor, suggesting a participation of KiSS1 in the Tachykinin receptor signaling pathway.
One of the strengths of our pilot study is the prospective recruitment of NSCLC cases and healthy subjects that guarantees the representativeness of the entire spectrum of cases encountered in the considered study timeframe, during the routine clinical practice in our institute. As regards the study limitation, despite the pilot nature of this investigation, our results are encouraging and suggest a possible role of KiSS1 as NSCLC biomarker. Larger prospective studies are needed to better explore and characterize the association between KiSS1 and disease status.
Conclusions
In the present translational study, we found an increase of KiSS1 levels in liquid biopsies of NSCLC patients versus healthy donors, a soluble KiSS1 release in media of NSCLC cells upon exposure to anticancer agents and apoptosis modulation by KiSS1-derived peptides. Our results highlight promising features of KiSS1 as a possible NSCLC biomarker, although further studies are needed to validate our clinical findings.
Acknowledgments
We thank all the patients taking part in this pilot study. We are grateful to Paola Suatoni, Federica Pirovano and Elena Bertocchi (Thoracic Surgery Unit) for their support in sample handling; and to Lara Girelli (Thoracic Surgery Unit) for discussion.
Funding: This work was supported by a grant from Fondazione CARIPLO-Regione Lombardia to PP (grant #2016-1019).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-52/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-52/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-52/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-52/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The research was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was obtained from all subjects upon approval of the study by institutional review board Fondazione IRCCS Istituto Nazionale dei Tumori (protocol number INT 167/16).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA Cancer J Clin 2022;72:7-33. [Crossref] [PubMed]
- Gandhi L, Rodríguez-Abreu D, Gadgeel S, et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:2078-92. [Crossref] [PubMed]
- Rottenberg S, Disler C, Perego P. The rediscovery of platinum-based cancer therapy. Nat Rev Cancer 2021;21:37-50. [Crossref] [PubMed]
- Mamdani H, Matosevic S, Khalid AB, et al. Immunotherapy in Lung Cancer: Current Landscape and Future Directions. Front Immunol 2022;13:823618. [Crossref] [PubMed]
- Chen Z, Fillmore CM, Hammerman PS, et al. Non-small-cell lung cancers: a hetero-geneous set of diseases. Nat Rev Cancer 2014;14:535-46. [Crossref] [PubMed]
- Segerman A, Niklasson M, Haglund C, et al. Clonal Variation in Drug and Radiation Response among Glioma-Initiating Cells Is Linked to Proneural-Mesenchymal Tran-sition. Cell Rep 2016;17:2994-3009. [Crossref] [PubMed]
- Bracht JWP, Mayo-de-Las-Casas C, Berenguer J, et al. The Present and Future of Liquid Biopsies in Non-Small Cell Lung Cancer: Combining Four Biosources for Diagnosis, Prognosis, Prediction, and Disease Monitoring. Curr Oncol Rep 2018;20:70. [Crossref] [PubMed]
- Nash KT, Phadke PA, Navenot JM, et al. Requirement of KISS1 secretion for multi-ple organ metastasis suppression and maintenance of tumor dormancy. J Natl Cancer Inst 2007;99:309-21. [Crossref] [PubMed]
- Kotani M, Detheux M, Vandenbogaerde A, et al. The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled re-ceptor GPR54. J Biol Chem 2001;276:34631-6. [Crossref] [PubMed]
- Harihar S, Pounds KM, Iwakuma T, et al. Furin is the major proprotein convertase required for KISS1-to-Kisspeptin processing. PLoS One 2014;9:e84958. [Crossref] [PubMed]
- Lee JH, Welch DR. Suppression of metastasis in human breast carcinoma MDA-MB-435 cells after transfection with the metastasis suppressor gene, KiSS-1. Cancer Res 1997;57:2384-7. [PubMed]
- Clarke H, Dhillo WS, Jayasena CN. Comprehensive Review on Kisspeptin and Its Role in Reproductive Disorders. Endocrinol Metab (Seoul) 2015;30:124-41. [Crossref] [PubMed]
- Guzman S, Brackstone M, Radovick S, et al. KISS1/KISS1R in Cancer: Friend or Foe? Front Endocrinol (Lausanne) 2018;9:437. [Crossref] [PubMed]
- Jiffar T, Yilmaz T, Lee J, et al. KiSS1 mediates platinum sensitivity and metastasis suppression in head and neck squamous cell carcinoma. Oncogene 2011;30:3163-73. [Crossref] [PubMed]
- Kim JN, Kim TH, Yoon JH, et al. Kisspeptin Inhibits Colorectal Cancer Cell Inva-siveness by Activating PKR and PP2A. Anticancer Res 2018;38:5791-8. [Crossref] [PubMed]
- Zuco V, Cassinelli G, Cossa G, et al. Targeting the invasive phenotype of cispla-tin-resistant non-small cell lung cancer cells by a novel histone deacetylase inhibitor. Biochem Pharmacol 2015;94:79-90. [Crossref] [PubMed]
- Corno C, Perego P. KiSS1 in regulation of metastasis and response to antitumor drugs. Drug Resist Updat 2019;42:12-21. [Crossref] [PubMed]
- Popa SM, Clifton DK, Steiner RA. The role of kisspeptins and GPR54 in the neuro-endocrine regulation of reproduction. Annu Rev Physiol 2008;70:213-38. [Crossref] [PubMed]
- Corno C, Gatti L, Arrighetti N, et al. Axl molecular targeting counteracts aggressive-ness but not platinum-resistance of ovarian carcinoma cells. Biochem Pharmacol 2017;136:40-50. [Crossref] [PubMed]
- Vescovi AL, Parati EA, Gritti A, et al. Isolation and cloning of multipotential stem cells from the embryonic human CNS and establishment of transplantable human neural stem cell lines by epigenetic stimulation. Exp Neurol 1999;156:71-83. [Crossref] [PubMed]
- Bez A, Corsini E, Curti D, et al. Neurosphere and neurosphere-forming cells: mor-phological and ultrastructural characterization. Brain Res 2003;993:18-29. [Crossref] [PubMed]
- Hosmer DW Jr, Lemeshow S. Applied Logistic Regression. Second Edition. New York, NY, USA: John Wiley & Sons, 2000.
- Hollander M, Wolfe DA. Nonparametric Statistical Methods. Second Edition. New York, NY, USA: John Wiley & Sons, 1999.
- Artusi R, Verderio P, Marubini E. Bravais-Pearson and Spearman correlation coeffi-cients: meaning, test of hypothesis and confidence interval. Int J Biol Markers 2002;17:148-51. [Crossref] [PubMed]
- Pastorino U, Boffi R, Marchianò A, et al. Stopping Smoking Reduces Mortality in Low-Dose Computed Tomography Screening Participants. J Thorac Oncol 2016;11:693-9. [Crossref] [PubMed]
- Jayasena CN, Comninos AN, Januszewski A, et al. Plasma kisspeptin: a potential biomarker of tumor metastasis in patients with ovarian carcinoma. Clin Chem 2012;58:1061-3. [Crossref] [PubMed]
- Dhillo WS, Chaudhri OB, Patterson M, et al. Kisspeptin-54 stimulates the hypotha-lamic-pituitary gonadal axis in human males. J Clin Endocrinol Metab 2005;90:6609-15. [Crossref] [PubMed]
- Horstmann M, Krause F, Steinbach D, et al. Evaluation of Plasmatic Kisspetin-10 as a Biomarker for Malignancy and Subtype Differentiation in Small Renal Tumours. Urol Int 2017;98:177-83. [Crossref] [PubMed]
- Ergen A, Canbay E, Bugra D, et al. Plasma Kisspeptin-54 levels in gastric cancer pa-tients. Int J Surg 2012;10:551-4. [Crossref] [PubMed]
- Canbay E, Ergen A, Bugra D, et al. Kisspeptin-54 levels are increased in patients with colorectal cancer. World J Surg 2012;36:2218-24. [Crossref] [PubMed]
- Brückl WM, Wirtz RM, Bertsch T, et al. Liquid Biopsy: Detection of Molecular Markers for Treatment Decisions in Lung Cancer. Pneumologie 2017;71:151-63. [PubMed]


