
Prognostic implications of cellular senescence in resected non-small cell lung cancer
Introduction
Non-small cell lung cancer (NSCLC) remains a devastating disease and the leading cause of cancer-related death in men and women worldwide (1,2). Approximately 85% of all lung malignancies are classified as NSCLC (3), for which the predicted 5-year relative survival rate for newly diagnosed cases with squamous and adenocarcinoma histology is only 21.7% and 28.5%, respectively (4). Adenocarcinoma is the most common histological subtype accounting for >50% of cases (5) followed by squamous histology representing 25% to 30% of NSCLC (6).
Although surgery provides for most patients with NSCLC the pathway to cure and long-term survival (7), about 30% to 60% of patients with completely resected early-stage NSCLC will ultimately develop local, regional and/or distant recurrence or a second-primary lung cancer (8). Resectable locally advanced NSCLC is often treated with platinum-based chemotherapy in neoadjuvant or adjuvant setting, depending on mediastinal nodal involvement (9). However, recurrence rate of resected stage IIIA NSCLC in the first and second year is 41% and 36%, respectively (10). Hence, cure and long-term survival for NSCLC remains hard to achieve.
Cellular senescence, an emerging hallmark of cancer (11), is a durable and irreversible cell cycle arrest with secretory features, macromolecular damage and altered metabolism that is elicited in response to different stresses (12) {such as (I) DNA damage (i.e., DNA damage-induced senescence); (II) the activation of oncogenes (i.e., oncogene-induced senescence); (III) various anticancer drugs [i.e., therapy-induced senescence (TIS)] (13)} and that often activates a persistent DNA-damage response (14). Due to the induction of a durable and generally irreversible cell cycle arrest, cellular senescence is often considered as an endogenous tumor suppressor mechanism (11,13). However, senescent malignant and non-malignant cells stay metabolically active and can secrete a plethora of largely pro-inflammatory cytokines, chemokines, growth factors and matrix-remodeling proteases—collectively known as the senescence-associated secretory phenotype (SASP). This SASP protects malignant cells from immune clearance and affects the surrounding tumor microenvironment (TME), ultimately stimulating cancer relapse and metastasis (15,16). Also, the stability of the senescence-associated cell-cycle arrest might be hindered by the genomic instability intrinsic to malignant cells (12,17), thus producing more aggressive variants (18) and boosting the capacity to drive tumor growth (19,20). As such, the role of cellular senescence in cancer is ambiguous (11).
Identification and quantification of senescent cells is a challenging task since currently, there are no specific and universal markers for senescent cells (12,14). Also, the detection of senescence-associated beta-galactosidase (SA-β-Gal) activity and often considered as the gold standard for identifying cells, is only applicable in fresh snap-frozen tissue samples (21). Therefore, detection of cellular senescence in archival formalin-fixed paraffin-embedded (FFPE) samples should be achieved by combining the measurements of different markers for cellular senescence, as previously reported (12,14). As a result, in vivo and ex vivo evidence of cellular senescence in cancer patient samples is sparse and is only now catching up (12).
In this context, we assessed the clinical impact and significance of the presence of cellular senescence in NSCLC patients, treated with or without stage-dependent (neo)adjuvant therapy. Surgically resected FFPE samples were stained using a combination of four immunohistochemical senescence markers as previously described (12,14), i.e., lipofuscin aggregates, cell cycle inhibitors p16INK4a and p21WAF1/Cip1 and proliferation marker Ki67. Correlation with clinicopathological parameters and outcomes, including and overall survival (OS) and disease-free survival (DFS) was calculated. We present the following article in accordance with the STROBE reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-192/rc).
Methods
Patient and tissue sampling
Primary tumor specimens, including clinicopathological parameters, were obtained from 155 NSCLC patients with postsurgical pathological tumor-node-metastasis (pTNM) stage I–IV (8th edition), treated with or without stage-dependent (neo)adjuvant therapy, who underwent surgery at the Antwerp University Hospital (UZA) between 2007 and 2020, and were made available by the Biobank Antwerp (reference number BB19049), Antwerp, Belgium; ID: BE 71030031000 (22) of the UZA. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of the Antwerp University Hospital (UZA) (reference number 19/18/236). Individual consent for this retrospective analysis was waived. OS was date of surgery until death by any cause. Patients without a date of death were censored at time of last follow-up. DFS was date of surgery until the first event of either recurrent disease or death.
Immunohistochemistry
FFPE tissue samples from 155 NSCLC patients were retrieved. Four separate, sequential and adjacent FFPE sections (5 µm thick) were cut using a HM340E microtome (Thermo Scientific®) and mounted on positively charged Superfrost Plus (Thermo Scientific®) slides that enable better adherence of the section, and stored at 4 ℃. Prior to staining, slides were placed in a 65 ℃ oven for 2 hours to melt the paraffin. Both positive and negative controls were included in all experiments.
Cellular senescence was assessed using a combination of four immunohistochemical senescence markers, i.e., lipofuscin aggregates, cell cycle inhibitor p16INK4a and p21WAF1/Cip1 and proliferation marker Ki67, as previously described (12,14), as the detection of SA-β-Gal activity is only applicable in fresh snap-frozen tissue samples (21).
Lipofuscin staining
The first step involved deparaffinization by washing in xylene for 8 minutes and rehydration for 5 minutes through graded isopropanol solutions of 100%, 95%, 80%, 70% and 50%, respectively. Slides were then washed in wash solution [phosphate buffered saline (PBS) + 0.3% triton X-100] and incubated for 1 hour with 1% bovine serum albumin at room temperature to block non-specific binding of antibody. Next, the slides were washed in wash solution and incubated for 5 minutes with 3.5% H2O2 (Acros Organics®) at room temperature to block endogenous peroxidase activity. Slides were then washed in wash solution, followed by 5 minutes of incubation in 50% and 70% EtOH, respectively. Then, filtered (Millex-GP® syringe filter unit, a 33 mm diameter filter with 0.22 µm pore size) SenTraGorTM (Arriani pharmaceuticals®), a biotinylated Sudan Black B based chemical reagent, was applied on the slides and covered with a cover slip to avoid evaporation during the following incubation of 10 minutes at 37 ℃. Afterwards, the coverslip of each slide was removed gently, and the slides were washed in 50% EtOH for 5 minutes at room temperature. Next, the slides were washed in PBS + 0.5% triton X-100 for 3 minutes at room temperature to increase permeabilization, and then incubated for 1 hour at 37 ℃ with the anti-biotin primary antibody (Abcam®, Ab201341) at a dilution of 1/200 and covered with a cover slip to avoid evaporation. Afterwards, the coverslip of each slide was removed gently, the slides were washed in wash solution and incubated with EnVision Flex+ Mouse LINKER (Dako®) for 15 minutes at room temperature to amplify the signal of the primary antibody. The slides were then washed in wash solution and incubated with Envision Flex/HRP (Dako®) for 45 minutes at room temperature. After the incubation, slides were washed in wash solution and EnVision Flex HRP Magenta Substrate Chromogen/Substrate Buffer (Dako®) was applied for 5 minutes at room temperature. The slides were washed in wash solution and counterstained with hematoxylin for 1 minute. Finally, the slides were dehydrated for 1 minute in isopropanol solutions of 70%, 95% and 100%, respectively, cleared in xylene for 2 minutes and mounted with Quick-D mounting medium (Klinipath®). A positive and negative control was included in each staining run and consisted of liver tissue and replacement of SenTraGorTM reagent by PBS, respectively.
p16INK4a, p21WAF1/Cip1 and Ki67
The first step involved deparaffinization by washing in xylene for 8 minutes and rehydration for 5 minutes through graded isopropanol solutions of 100%, 95% and 70%, respectively, followed by 5 minutes in distilled water. Subsequently, antigen retrieval with target retrieval solution—high pH (Dako®) in a PT Link pre-treatment module (Dako®) was conducted at pH 9.0 for 15 minutes (p21WAF1/Cip1) or 20 minutes (p16INK4a and Ki67) at 97 ℃. Slides were washed for 5 minutes in Envision Flex Wash Buffer (Dako®) between each step. Endogenous peroxidase activity was quenched by incubating the slides in peroxidase blocking reagent for 5 minutes. Incubation with primary mouse monoclonal anti-human p16INK4a antibody (clone E6H4, ready to use, Roche Diagnostics®), p21WAF1/Cip1 antibody (clone SX118, dilution 1/100, Agilent Technologies®) and Ki67 antibody (clone MIB-1, ready to use, Agilent Technologies®) was performed for 30, 40 and 20 minutes, respectively, at room temperature. Primary antibody incubation was followed by incubation with ready to use visualization reagent for 30 minutes (p16INK4a) or mouse enhanced polymer-based linker (Mouse Linker, Dako®) for 15 minutes (p21WAF1/Cip1 and Ki67). Detection and signal visualization was performed using visualization reagent (polymer reagent conjugated with horseradish peroxidase and affinity purified goat anti-mouse Fab’ antibody fragments, supplied in stabilizing solution comprising preservatives and stabilizing protein, CINtec® Histology Kit) (p16INK4a) or Envision FLEX/HRP (ready to use, 20 minutes, Dako®) (p21WAF1/Cip1 and Ki67) followed by 10 minutes incubation with DAB Substrate-Chromogen solution (CINtec® Histology Kit) (p16INK4a), and 5 minutes (p21WAF1/Cip1) or 10 minutes (Ki67) incubation with the Dako Liquid DAB+ Substrate Chromogen System (Dako®). The slides were counterstained with hematoxylin for 2 minutes. Finally, the slides were dehydrated for 1 minute in isopropanol solutions of 70%, 95% and 100%, respectively, cleared in xylene for 2 minutes and mounted with Quick-D mounting medium (Klinipath®). A positive and negative control was included in each staining run and consisted for (I) p16INK4a respectively of tonsil tissue and replacement of primary mouse monoclonal anti-human p16INK4a antibody by Negative Reagent Control (Roche Diagnostics®), according to the manufacturer’s instruction; for (II) p21WAF1/Cip1 respectively of squamous cell lung carcinoma tissue and replacement of primary mouse monoclonal anti-human p21WAF1/Cip1 antibody by FLEX Negative Control Mouse (Dako®), according to the manufacturer’s instruction and for (III) Ki67 respectively of tonsil tissue and replacement of primary mouse monoclonal anti-human Ki67 antibody by FLEX Negative Control Mouse (Dako®), according to the manufacturer’s instruction.
Assessment of immunohistochemical markers
Slides were scanned using an Ultra Fast (digital pathology slide scanner, Philips®) and images were analyzed using Image Management System viewer (pathology case viewer, Philips®). The percentage of tumor cells expressing the examined senescence markers was assessed in all optical ×200 fields available and the mean score was used to score each case. Lipofuscin accumulation, p16INK4a and p21WAF1/Cip1 protein expression, and Ki67 expression were detected as cytoplasmic, mixed cytoplasmic/nuclear and nuclear stainings, respectively. Both an independent observer (AD) and a pathologist (VS) scored the different slides and scoring was performed blinded for clinical data.
A senescence signature (SS) was defined by the presence of high-level lipofuscin and high p16INK4a and/or p21WAF1/Cip1 expression (≥30% NSCLC cells positive) in combination with low Ki67 expression (<30% NSCLC cells positive). An SS-p16 was defined by the presence of high-level lipofuscin and solely high p16INK4a expression (≥30% NSCLC cells positive) in combination with low Ki67 expression (<30% NSCLC cells positive). An SS-p21 was defined by the presence of high-level lipofuscin and solely high p21WAF1/Cip1 expression (≥30% NSCLC cells positive) in combination with low Ki67 expression (<30% NSCLC cells positive). An SS-p16-p21 was defined by the presence of high-level lipofuscin and high p16INK4a and p21WAF1/Cip1 expression (≥30% NSCLC cells positive) in combination with low Ki67 expression (<30% NSCLC cells positive).
Mutation analysis
Epidermal growth factor receptor (EGFR) and Kirsten rat sarcoma virus (KRAS) mutation status was determined as part of routine clinical practice by high-resolution melting analysis or next generation sequencing. TP53 mutation status was determined in a subset of patients as previously described (23).
Statistical analysis
The statistical analysis was performed using IBM SPSS Statistics version 28.0. Correlation between individual senescence markers was analyzed using Spearman correlation. Association between SS and OS and DFS was studied using simple and multiple Cox proportional hazards (Cox PH) model to adjust for covariates (i.e., age, pTNM stage I–III and adjuvant therapy). Survival (OS) and recurrence (DFS) curves were estimated using the Kaplan-Meier (KM) method. Differences between patients with and without an SS were studied using independent samples t-test or Mann-Whitney U test for continuous variables and Chi-square test or Fisher’s exact test for categorical variables. P values <0.05 were considered to be statistically significant, and all tests were two-sided. Graphical presentation was performed using GraphPad Prism version 9.2.0.
Results
Presence of an SS, as assessed by immunohistochemical staining
The baseline patient, tumor and treatment characteristics of the total patient cohort and the homogenous patient cohort used for distribution analyses with clinicopathological parameters and survival analyses, respectively, are summarized in Table 1. The immunohistochemical expression of the four senescence markers in the total patient cohort and according to the tumoral SS status (i.e., SS and no SS) is summarized in Table 2. Figure 1 shows typical immunohistochemical correlation of high-level lipofuscin accumulation, high p16INK4a and p21WAF1/Cip1 expression and low Ki67 expression, representing an SS. The presence of an SS was reported for 48 out of 155 NSCLC patients.
Table 1
| Characteristic | Total cohort (n=155) | Homogenous cohort (n=100) |
|---|---|---|
| Age at diagnosis, years | ||
| Mean (95% CI) | 65 (64–66) | 66 (64–68) |
| Median (min–max) | 65 (39–85) | 66 (45–85) |
| Gender, n (%) | ||
| Female | 58 (37.4) | 40 (40.0) |
| Male | 97 (62.6) | 60 (60.0) |
| Smoking history, n (%) | ||
| Positive | 140 (90.3) | 92 (92.0) |
| Negative | 15 (9.7) | 8 (8.0) |
| Histology, n (%) | ||
| Adenocarcinoma | 139 (89.7) | 100 (100.0) |
| Squamous cell carcinoma | 14 (9.0) | NA |
| Adenosquamous | 1 (0.6) | NA |
| NOS | 1 (0.6) | NA |
| Grade of differentiation, n (%) | ||
| Well differentiated | 50 (32.3) | 40 (40.0) |
| Moderately differentiated | 56 (36.1) | 37 (37.0) |
| Poorly differentiated | 49 (31.6) | 23 (23.0) |
| Tumor size, cm, n (%) | ||
| 0–1.9 | 35 (22.6) | 29 (29.0) |
| 2–3.9 | 67 (43.2) | 48 (48.0) |
| 4–5.9 | 31 (20.0) | 12 (12.0) |
| ≥6 | 22 (14.2) | 11 (11.0) |
| pTNM stage (8th edition), n (%) | ||
| Stage I | 67 (43.2) | 54 (54.0) |
| Stage II | 41 (26.5) | 21 (21.0) |
| Stage III | 39 (25.2) | 25 (25.0) |
| Stage IV | 8 (5.2) | NA |
| Surgical resection, n (%) | ||
| R0 | 146 (94.2) | 100 (100.0) |
| R1 | 9 (5.8) | NA |
| Mutation, positive/tested | ||
| p53 | 19/53 | 12/36 |
| EGFR | 10/107 | NA |
| KRAS | 28/63 | 23/40 |
| Treatment | ||
| Neoadjuvant therapy, n (%) | 39 (25.2) | NA |
| Cisplatin- or carboplatin-based doublet chemotherapy | 36 (23.2) | NA |
| Cisplatin- or carboplatin-based doublet with radiotherapy | 2 (1.3) | NA |
| Pembrolizumab with radiotherapy | 1 (0.6) | NA |
| Surgery, n (%) | 155 (100.0) | 100 (100.0) |
| Adjuvant therapy, n (%) | 46 (29.7) | 27 (27.0) |
| Cisplatin- or carboplatin-based chemotherapy | 29 (18.7) | 21 (21.0) |
| Cisplatin- or carboplatin-based chemotherapy with radiotherapy | 10 (6.5) | 6 (6.0) |
| Radiotherapy only | 7 (4.5) | NA |
CI, confidence interval; NA, not applicable; NOS, not otherwise specified; pTNM, pathological tumor-node-metastasis; EGFR, epidermal growth factor receptor; KRAS, Kirsten rat sarcoma virus.
Table 2
| Senescence marker | Range | Median | Mean ± SD | High expression, n (%) | Low expression, n (%) | Negative expression, n (%) |
|---|---|---|---|---|---|---|
| % Lipofuscin + cells | 0–100 | 50.0 | 46.9±30.5 | 103 (66.5) | 52 (33.5) | 8 (5.2) |
| SS (n=48) | 30–100 | 67.5 | 65.3±19.7 | 48 (100.0) | 0 | 0 |
| No SS (n=107) | 0–90 | 30.0 | 38.7±31.0 | 55 (51.4) | 52 (48.6) | 8 (7.5) |
| % p16INK4a + cells | 0–100 | 30.0 | 38.5±38.3 | 78 (50.3) | 77 (49.7) | 41 (26.5) |
| SS (n=48) | 0–100 | 62.5 | 58.8±33.8 | 39 (81.3) | 9 (18.8) | 5 (10.4) |
| No SS (n=107) | 0–100 | 10.0 | 29.4±36.8 | 39 (36.4) | 68 (63.6) | 36 (33.6) |
| % p21WAF1/Cip1 + cells | 0–90 | 20.0 | 28.1±26.9 | 61 (39.4) | 94 (60.6) | 14 (9.0) |
| SS (n=48) | 0–90 | 30.0 | 38.2±28.9 | 25 (52.1) | 23 (47.9) | 2 (4.2) |
| No SS (n=107) | 0–90 | 15.0 | 23.5±24.8 | 36 (33.6) | 71 (66.4) | 12 (11.2) |
| % Ki67 + cells | 0–95 | 17.5 | 28.2±25.4 | 60 (38.7) | 95 (61.3) | 3 (1.9) |
| SS (n=48) | 0–28 | 8.3 | 10.2±7.9 | 0 | 48 (100.0) | 2 (4.2) |
| No SS (n=107) | 0–95 | 35.0 | 36.2±26.4 | 60 (56.1) | 47 (43.9) | 1 (0.9) |
High expression: ≥30% NSCLC cells positive; low expression: <30% NSCLC cells positive. SS, senescence signature; SD, standard deviation; NSCLC, non-small cell lung cancer.
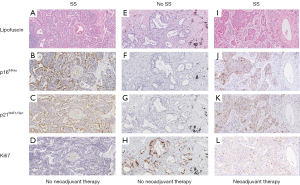
Correlation of individual senescence markers and according to tumoral SS status
First, we investigated whether the expression of the four individual senescence markers in the total patient cohort (n=155) and according to the tumoral SS status were correlated to each other to assess whether the individual senescence markers are interdependent for immunohistochemical detection of cellular senescence. Immunohistochemical p16INK4a expression, but not lipofuscin accumulation or p21WAF1/Cip1 expression, was significantly inversely correlated with Ki67 expression (correlation coefficient =−0.32, P value <0.001). Immunohistochemical lipofuscin accumulation and p21WAF1/Cip1 expression did not correlate to any of the individual senescence markers.
In patients with a tumoral SS (n=48), immunohistochemical p16INK4a expression was positively correlated with high-level lipofuscin accumulation (<30% NSCLC cells positive) (correlation coefficient =0.26, P value =0.071) and significantly inversely correlated with p21WAF1/Cip1 expression (correlation coefficient =−0.34, P value =0.018). Conversely, in patients with no tumoral SS (n=107), immunohistochemical p16INK4a expression was significantly inversely correlated with lipofuscin accumulation (correlation coefficient =−0.24, P value =0.014) and significantly positively correlated with p21WAF1/Cip1 expression (correlation coefficient =0.22, P value =0.024). Also, in patients with no tumoral SS, immunohistochemical Ki67 expression was significantly positively correlated with lipofuscin accumulation (correlation coefficient =0.33, P value <0.001) and significantly inversely correlated with p16INK4a expression (correlation coefficient =−0.22, P value =0.023).
Taken together, these results demonstrate an inverse correlation between immunohistochemical p16INK4a expression and Ki67 expression irrespective of an SS. According to the tumoral SS status, p16INK4a expression demonstrate differential correlations with lipofuscin accumulation and p21WAF1/Cip1 expression.
Correlation of an SS with clinicopathological parameters
Next, we investigated whether the presence of an SS was associated with clinicopathological parameters in the total patient cohort of 155 patients. Presence of an SS was not significantly associated with clinical parameters (age, gender and smoking history), pathological parameters (grade of differentiation, tumor size and pTNM stage) or EGFR and KRAS mutation status. However, an SS was significantly more present in case of confirmed p53 wild-type samples compared to samples with a confirmed p53 mutation (50.0% versus 21.1%, P=0.039).
As commonly used cancer interventions have been associated with the induction of cellular senescence (i.e., TIS) in cancer cells (16), we next analyzed the expression of the individual senescence markers as well as the presence of an SS according to neoadjuvant therapy. Patients who received neoadjuvant therapy showed a significantly higher mean lipofuscin accumulation (59.1% versus 42.8%, P=0.004) and Ki67 expression (37.4% versus 25.1%, P=0.015) compared to patients who did not receive any neoadjuvant treatment. p16INK4a (36.5% versus 39.2%, P=0.70) and p21WAF1/Cip1 (29.6% versus 27.5%, P=0.68) expression were very comparable between patients with or without neoadjuvant therapy. Regarding the presence of an SS according to neoadjuvant therapy, an SS was not significantly more present in patients with neoadjuvant therapy compared to patients without neoadjuvant therapy (28.2% versus 31.9%, P=0.67). However, in case an SS was present, patients who did receive neoadjuvant therapy showed a higher mean lipofuscin accumulation (73.6% versus 62.7%, P=0.11) and a significantly higher p16INK4a expression (77.3% versus 53.4%, P=0.038) compared to patients without neoadjuvant therapy. p21WAF1/Cip1 expression (39.1% versus 38.0%, P=0.91) and Ki67 expression (9.5% versus 10.4%, P=0.75) were very comparable between patients with or without neoadjuvant therapy.
In patients who did receive neoadjuvant therapy (n=39), an SS was more present in adenocarcinoma samples (9/24, 37.5%) compared to samples with another histological subtype (2/15, 13.3%). Also, within this subpopulation of which the p53 mutation status was determined (n=10), an SS was more present in p53 wild-type samples (3/4, 75.0%) compared to samples which harbored a confirmed p53 mutation (1/6, 16.7%). However, the analyses were performed in a small sample size and differences were non-significant.
Overall, these above-mentioned observations indicate that an SS is more present in p53 wild-type samples, and neoadjuvant therapy might induce a new SS that is more pronounced or reinforce a pre-existing SS by increasing lipofuscin accumulation and p16INK4a expression.
Correlation between the presence of SS and survival
We next analyzed the relationship between the presence of an SS and OS in a histologically heterogenous patient cohort of 139 patients who had surgery with curative intent, i.e., excluding pTNM stage IV and resections with microscopically residual tumor (R1). With a median follow-up of 53 months, KM OS analysis showed a difference in median OS for patients with an SS compared to patients without an SS with a clear trend for worse OS for patients with an SS approaching significance (62 versus 88 months, P=0.059) (Figure 2A). Next, we selected a more histologically homogenous patient group consisting of 100 patients with pTNM stages I–III, according to the 8th edition, who fulfilled the following criteria: (I) one type of histology, i.e., adenocarcinoma, (II) without known EGFR mutation, (III) curative (R0) resection and (IV) no neoadjuvant systemic therapy or radiotherapy as neoadjuvant therapy is capable of inducing TIS (16) (Table 1). In this histologically homogenous patient cohort with a median follow-up of 57 months, KM OS analysis showed a significant difference in median OS for patients with an SS compared to patients without an SS (53 versus 141 months, P=0.005) (Figure 2B) with a 5-year OS rate of 44.9% versus 66.9%, respectively. The presence of an SS served as a significant prognostic factor for OS in simple [n=100; hazard ratio (HR) =2.17; P=0.007] and multiple (n=100; HR =2.03; P=0.014) Cox PH model analysis correcting for age, pTNM stage I–III and adjuvant therapy (Table 3). Interestingly, no individual senescence marker served as a significant prognostic factor for OS in simple and multiple Cox PH model analysis when scored as continuous or categorical [high (≥30% NSCLC cells positive)/low (<30% NSCLC cells positive) expression] variables. Regarding DFS, KM analysis showed difference in median DFS for patients with an SS compared to patients without an SS (45 versus 55 months) with a trend towards significance (P=0.25) (Figure 2C).
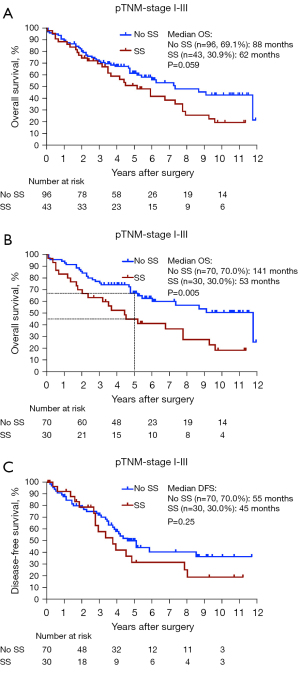
Table 3
| Characteristics | HR | 95% CI | P value |
|---|---|---|---|
| Simple Cox PH model analysis | |||
| SS | 2.17 | 1.24–3.80 | 0.007* |
| SS-p16 | 2.23 | 1.05–4.73 | 0.037* |
| SS-p21 | 1.43 | 0.50–4.07 | 0.51 |
| SS-p16-p21 | 2.74 | 1.29–5.81 | 0.009* |
| Multiple Cox PH model analysis correcting for age, pTNM stage I–III and adjuvant therapy | |||
| SS | 2.03 | 1.15–3.57 | 0.014* |
| SS-p16 | 2.10 | 0.97–4.53 | 0.059 |
| SS-p21 | 1.31 | 0.46–3.79 | 0.61 |
| SS-p16-p21 | 2.55 | 1.19–5.45 | 0.016* |
*, HR is significant. Cox PH; Cox proportional hazards; HR, hazard ratio; CI, confidence interval; SS, senescence signature; pTNM, pathological tumor-node-metastasis.
Collectively, these results demonstrate that the presence of an SS has significant prognostic implications in resected NSCLC adenocarcinoma.
p16INK4a and p21WAF1/Cip1 are both cell cycle inhibitor proteins, however, each capable of inducing senescence via different downstream pathways (24) and show independent immunohistochemical expression as mentioned above. Therefore, we determined the prognostic value of an SS defined by high-level lipofuscin plus low Ki67 expression, combined with (I) solely high p16INK4a expression (≥30% NSCLC cells positive, i.e., SS-p16); (II) solely high p21WAF1/Cip1 expression (≥30% NSCLC cells positive, i.e., SS-p21); and (III) both high p16INK4a and high p21WAF1/Cip1 (≥30% NSCLC cells positive, i.e., SS-p16-p21) expression. Compared to patients without any SS phenotype, KM OS analysis showed a worse OS for patients with an SS-p16 (53 versus 141 months, P=0.032) as well as for patients with an SS-p21 (115 versus 141 months, P=0.51), and patients with an SS-p16-p21 (44 versus 141 months, P=0.006) (Figure 3). The presence of an SS-p16 and an SS-p16-p21, but not an SS-p21, served as a significant prognostic factor for OS in simple Cox PH model analysis (Table 3). In multiple Cox PH model analysis correcting for age, pTNM stage I–III and adjuvant therapy, only the presence of an SS-p16-p21 remained as a significant prognostic factor whereas the presence of an SS-p16 and an SS-p21 did not (Table 3 and Figure 4).
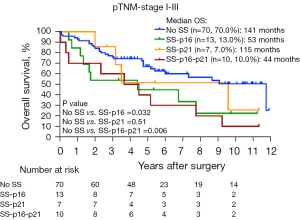
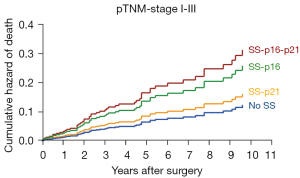
These results thus indicate that the presence of an SS particularly based on high p16INK4a expression has more profound impact on OS compared to an SS based on solely high p21WAF1/Cip1 expression.
Discussion
Cellular senescence is considered as a protective mechanism, in addition to programmed cell death, for maintaining tissue homeostasis by inducing an irreversible proliferative arrest in dysfunctional and diseased cells (11), and has beneficial functions in a variety of physiological and pathological processes, such as embryogenesis and wound healing (25). In cancer, cellular senescence has long been considered as an endogenous tumor suppressor mechanism by inducing senescence in pre- or fully malignant cells, thereby limiting malignant progression (11). However, despite the clear benefit of the senescence-associated growth arrest for preventing the expansion of pre- or fully malignant cells, senescent malignant (26) and non-malignant cells (27) are capable of paradoxically stimulating tumor relapse, progression and metastasis. This is presumably caused by the SASP as well as the genomic instability of malignant cells, overriding the senescence-associated cell cycle arrest over time (11).
Due to its importance in regulating various biological processes, and its ambiguous and even controversial role in cancer, in vivo evidence for cellular senescence in human tissue has gained more attention in the last decade (12). As a result, cellular senescence has recently been promoted as an emerging hallmark of cancer (11). However, little is known about the consequence of cellular senescence on prognosis and survival in distinct types of cancer. In breast cancer, high expression of a cellular senescence gene panel was associated with increased survival (28). In contrast, in hepatocellular carcinoma, a cellular senescence-associated gene signature in the TME was associated with worse DFS and OS (29). In prostate cancer, tumor-associated senescence has a dual role with tumor-suppressive as well as tumor-promoting properties, depending on deletion of the metalloproteinase inhibitor TIMP1 (26).
In NSCLC, only two previous reports demonstrated a negative prognostic value of individual immunohistochemical senescence markers, such as lipofuscin accumulation (30) and high p21WAF1/Cip1 and high Ki67 (31), however, both in rather histological heterogenous patient populations. On the contrary, loss of p16INK4a expression (<10%), which is a common feature especially in squamous cell carcinoma compared to adenocarcinoma (87% versus 50%, respectively), was associated with significantly worse OS (32). Nonetheless, to identify senescent cells with the highest accuracy, the International Cell Senescence Association (12) and Kohli et al. (14) recommend combining individual markers, as senescent cells lack specific and universal markers.
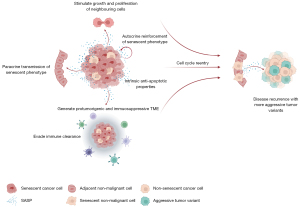
The present study demonstrates for the first time that the presence of a tumoral SS, based on the combination of four cellular senescence markers, negatively affects OS and DFS in NSCLC adenocarcinoma. In this context, senescent cancer cells could initially serve as a temporarily indolent cancer cell reservoir, with anti-apoptotic properties that evades immune clearance and molds a favorable surrounding TME by secreting SASP immunosuppressive cytokines and matrix-remodeling proteases. In later stages, the SASP can reinforce the senescent phenotype in an autocrine way, and paracrinally spread the senescent phenotype to adjacent malignant and non-malignant cells. With an increasing tumoral senescence burden, the accumulation of SASP can subsequently stimulate growth and proliferation of neighboring benign, premalignant and malignant cells (33). Finally, due to their genomic instability, and the possibility to acquire additional mutations that affect the function of cell-cycle arrest genes, senescent cancer cells can escape from the proliferative stall, probably resulting in loss of p16INK4a expression and compatible with previous findings of Sterlacci et al. (32), ultimately leading to disease recurrence and worse OS. The senescent phenotype spread by the tumoral SS to the adjacent non-tumoral tissue can contribute to the protumorigenic effects exhibited by the tumoral SS, ultimately promoting cancer relapse (27) (Figure 5). Of note, samples without a tumoral SS do not necessarily exclusively harbor non-senescent proliferating cancer cells but nevertheless can also harbor few senescent cancer cells. Since senescence is considered antagonistically pleiotropic (34), it could be suggested that the quantity of senescent cancer cells in these samples is not sufficient to exhibit similar protumorigenic effects as in samples with a high tumoral senescence burden, and the occasional senescent cells act primarily tumor suppressive through the senescent-associated cell cycle arrest. The relationship between the presence of a tumoral SS and DFS was less clear. Possibly, a tumoral SS does not necessarily result in a shorter DFS but rather, in case of disease recurrence, results in a more aggressive disease course due to more aggressive tumor variants produced by the tumoral SS (Figure 5). However, since the retrospective nature of the study, DFS was not an ideal primary endpoint.
Interestingly, Lin et al. (35) recently demonstrated that NSCLC adenocarcinoma patients with a highly expressed senescence-related gene signature experienced a significantly shorter OS compared to patients with low expression of these senescence-related genes, confirming our results, however, at transcriptional level.
Cellular senescence is predominantly regulated by the activation of cell cycle inhibitors p21WAF1/Cip1 and/or p16INK4a, as most senescence-inducing stresses leads to the activation of the cell cycle inhibitor pathways p53/p21WAF1/Cip1 and/or p16INK4a (24). Activation p21WAF1/Cip1 and/or p16INK4a will induce a stable cell cycle arrest by inhibition of cyclin-dependent kinase 1 (CDK1), CDK2, CDK4 and CDK6 and thereby prevents phosphorylation of the retinoblastoma protein (24), a tumor suppressor protein blocking S-phase entry (36). However, p21WAF1/Cip1 expression occurs earlier after senescence induction and is reversible upon tumor suppressor protein p53 inactivation, in contrast to p16INK4a that is frequently expressed late after senescence induction and irreversible upon p53 inactivation (14,37,38). As such, p53/p21WAF1/Cip1 pathway activation seems to be more involved in the initiation of senescence, while p16INK4a seem to be more crucial for maintaining the senescence-associated arrest (38). Interestingly, our data shows that an SS based on solely p16INK4a expression (i.e., SS-p16) and an SS based on high expression of both p16INK4a and high p21WAF1/Cip1 (i.e., SS-p16-p21) had a more profound impact on OS in contrast to an SS based on solely high p21WAF1/Cip1 expression (i.e., SS-p21). Possibly, an SS based on high p16INK4a expression is more durable and therefore more protumorigenic with established SASP production compared to a premature SS based on solely high p21WAF1/Cip1 expression.
Although cell cycle inhibitor pathways p53/p21WAF1/Cip1 and p16INK4a are intertwined, DNA-damage, oncogene activation and tumor suppressor inactivation mainly lead to the activation of tumor suppressor protein p53 and consequently p21WAF1/Cip1 through the Ras-Raf-MEK-ERK or AKT signaling pathways (24). In accordance, an SS was more present in p53 wild-type samples compared to p53 mutated samples, where induction of senescence via p53/p21WAF1/Cip1 pathway activation is presumably defective (23). Nonetheless, cellular senescence can also be induced through activation of p16INK4a, irrespective of p53/p21WAF1/Cip1 pathway activation (24).
Regarding TIS, an SS was not more present in patients who received neoadjuvant therapy compared to patients who did not receive any neoadjuvant therapy. However, the SS in patients who received neoadjuvant therapy was more pronounced based on the lipofuscin accumulation and p16INK4a expression, suggesting stronger induction of a new SS or reinforcement of a pre-existing SS by neoadjuvant therapy. Also, neoadjuvant therapy was more prone to induce an SS in adenocarcinoma and p53 wild-type samples compared to samples with other histology or p53 mutation. However, the results of these sub-analyses were non-significant and need to be interpreted with caution due to the small sample size.
This study has several limitations. The data represent a retrospective review of a cohort of patients seen at a single academic institution over a period of 14 years, and not all patients received their follow-up at our center. Therefore, OS was used as a primary endpoint.
Another limitation of the study, as mentioned above, is the sample size. However, even in this small population of NSCLC patients, significant differences in survival analyses were seen, suggesting that the presence of an SS could have a profound impact on OS. Nonetheless, our preliminary results need to be confirmed in a large-scale population and prospective study design.
In the future, we plan to enlarge our sample size and include more NSCLC patients with histological subtypes other than adenocarcinoma to better delineate the differences that could exist within histology and within EGFR and KRAS mutation status.
In conclusion, the presence of an SS particularly based on high p16INK4a expression was significantly negatively associated with OS in NSCLC adenocarcinoma after correction for age, pTNM stage and adjuvant therapy. Since senolytics (i.e., compounds that selectively target senescent cells) are currently being evaluated in clinical trials for age-related pathologies such as osteoarthritis (NCT04815902) and Alzheimer’s disease (NCT04063124) and are proposed as a novel cancer therapy (16,39-41), we suggest that adjuvant senolytic therapy could be an interesting strategy for NSCLC adenocarcinoma patients harboring an SS, ultimately to improve survival of these patients.
Acknowledgments
We thank Kristien Wouters [UZA, Edegem (Antwerp), Belgium] for assisting the statistical analyses.
This research was funded by donations from different donors, including Dedert Schilde vzw and Willy Floren.
Figure 5 was created with Biorender.com.
Funding: This work was supported by the University Research Fund (BOF) of the University of Antwerp, Antwerp, Belgium (No. FFB180188 to AD).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-192/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-192/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-192/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-192/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Ethics Committee of the Antwerp University Hospital (UZA) (reference number 19/18/236). Individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Planchard D, Popat S, Kerr K, et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2018;29:iv192-237. [Crossref]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Howlader N, Noone AM, Krapcho M, et al. Lung and Bronchus SEER 5-Year Relative Survival Rates, 2011-2017. Available online: https://seer.cancer.gov/explorer/application.html?site=47&data_type=4&graph_type=5&compareBy=subtype&chk_subtype_612=612&chk_subtype_613=613&chk_subtype_611=611&chk_subtype_610=610&series=9&sex=1&race=1&age_range=1&stage=101&advopt_precision=1&advopt_show_ci=on&advopt_display=2
- Girard N, Sima CS, Jackman DM, et al. Nomogram to predict the presence of EGFR activating mutation in lung adenocarcinoma. European Respiratory Journal 2012;39:366-72. [Crossref] [PubMed]
- Socinski MA, Obasaju C, Gandara D, et al. Clinicopathologic Features of Advanced Squamous NSCLC. J Thorac Oncol 2016;11:1411-22. [Crossref] [PubMed]
- Osarogiagbon RU, Veronesi G, Fang W, et al. Early-Stage NSCLC: Advances in Thoracic Oncology 2018. J Thorac Oncol 2019;14:968-78. [Crossref] [PubMed]
- Conforti F, Pala L, Pagan E, et al. Effectiveness of intensive clinical and radiological follow-up in patients with surgically resected NSCLC. Analysis of 2661 patients from the prospective MAGRIT trial. Eur J Cancer 2020;125:94-103. [Crossref] [PubMed]
- Postmus PE, Kerr KM, Oudkerk M, et al. Early and locally advanced non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2017;28:iv1-21. [Crossref] [PubMed]
- Lou F, Sima CS, Rusch VW, et al. Differences in patterns of recurrence in early-stage versus locally advanced non-small cell lung cancer. Ann Thorac Surg 2014;98:1755-60; discussion 1760-1. [Crossref] [PubMed]
- Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov 2022;12:31-46. [Crossref] [PubMed]
- Gorgoulis V, Adams PD, Alimonti A, et al. Cellular Senescence: Defining a Path Forward. Cell 2019;179:813-27. [Crossref] [PubMed]
- Hernandez-Segura A, Nehme J, Demaria M. Hallmarks of Cellular Senescence. Trends Cell Biol 2018;28:436-53. [Crossref] [PubMed]
- Kohli J, Wang B, Brandenburg SM, et al. Algorithmic assessment of cellular senescence in experimental and clinical specimens. Nat Protoc 2021;16:2471-98. [Crossref] [PubMed]
- Faget DV, Ren Q, Stewart SA. Unmasking senescence: context-dependent effects of SASP in cancer. Nat Rev Cancer 2019;19:439-53. [Crossref] [PubMed]
- Wang B, Kohli J, Demaria M. Senescent Cells in Cancer Therapy: Friends or Foes? Trends Cancer 2020;6:838-57. [Crossref] [PubMed]
- Roberson RS, Kussick SJ, Vallieres E, et al. Escape from therapy-induced accelerated cellular senescence in p53-null lung cancer cells and in human lung cancers. Cancer Res 2005;65:2795-803. [Crossref] [PubMed]
- Yang L, Fang J, Chen J. Tumor cell senescence response produces aggressive variants. Cell Death Discov 2017;3:17049. [Crossref] [PubMed]
- Medema JP. Escape from senescence boosts tumour growth. Nature 2018;553:37-8. [Crossref]
- Milanovic M, Fan DNY, Belenki D, et al. Senescence-associated reprogramming promotes cancer stemness. Nature 2018;553:96-100. [Crossref] [PubMed]
- Evangelou K, Lougiakis N, Rizou SV, et al. Robust, universal biomarker assay to detect senescent cells in biological specimens. Aging Cell 2017;16:192-7. [Crossref] [PubMed]
- BE 71030031000; Biobank Antwerp [BB19049], BBMR-ERIC, Belgian [BIORESOURCE].
- Deben C, Deschoolmeester V, Lardon F, et al. TP53 and MDM2 genetic alterations in non-small cell lung cancer: Evaluating their prognostic and predictive value. Crit Rev Oncol Hematol 2016;99:63-73. [Crossref] [PubMed]
- Paez-Ribes M, González-Gualda E, Doherty GJ, et al. Targeting senescent cells in translational medicine. EMBO Mol Med 2019;11:e10234. [Crossref] [PubMed]
- He S, Sharpless NE. Senescence in Health and Disease. Cell 2017;169:1000-11. [Crossref] [PubMed]
- Guccini I, Revandkar A, D’Ambrosio M, et al. Senescence Reprogramming by TIMP1 Deficiency Promotes Prostate Cancer Metastasis. Cancer Cell 2021;39:68-82.e9. [Crossref] [PubMed]
- Demaria M, O’Leary MN, Chang J, et al. Cellular Senescence Promotes Adverse Effects of Chemotherapy and Cancer Relapse. Cancer Discov 2017;7:165-76. [Crossref] [PubMed]
- Althubiti M, Lezina L, Carrera S, et al. Characterization of novel markers of senescence and their prognostic potential in cancer. Cell Death Dis 2014;5:e1528. [Crossref] [PubMed]
- Eggert T, Wolter K, Ji J, et al. Distinct Functions of Senescence-Associated Immune Responses in Liver Tumor Surveillance and Tumor Progression. Cancer Cell 2016;30:533-47. [Crossref] [PubMed]
- Giatromanolaki A, Kouroupi M, Balaska K, et al. A Novel Lipofuscin-detecting Marker of Senescence Relates With Hypoxia, Dysregulated Autophagy and With Poor Prognosis in Non-small-cell-lung Cancer. In Vivo 2020;34:3187-93. [Crossref] [PubMed]
- Dosaka-Akita H, Hommura F, Mishina T, et al. A risk-stratification model of non-small cell lung cancers using cyclin E, Ki-67, and ras p21: different roles of G1 cyclins in cell proliferation and prognosis. Cancer Res 2001;61:2500-4. [PubMed]
- Sterlacci W, Tzankov A, Veits L, et al. A comprehensive analysis of p16 expression, gene status, and promoter hypermethylation in surgically resected non-small cell lung carcinomas. J Thorac Oncol 2011;6:1649-57. [Crossref] [PubMed]
- Gonzalez-Meljem JM, Apps JR, Fraser HC, et al. Paracrine roles of cellular senescence in promoting tumourigenesis. Br J Cancer 2018;118:1283-8. [Crossref] [PubMed]
- Campisi J. Aging, tumor suppression and cancer: high wire-act! Mech Ageing Dev 2005;126:51-8. [Crossref] [PubMed]
- Lin W, Wang X, Wang Z, et al. Comprehensive Analysis Uncovers Prognostic and Immunogenic Characteristics of Cellular Senescence for Lung Adenocarcinoma. Front Cell Dev Biol 2021;9:780461. [Crossref] [PubMed]
- Giacinti C, Giordano A. RB and cell cycle progression. Oncogene 2006;25:5220-7. [Crossref] [PubMed]
- Alcorta DA, Xiong Y, Phelps D, et al. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci U S A 1996;93:13742-7. [Crossref] [PubMed]
- Beauséjour CM, Krtolica A, Galimi F, et al. Reversal of human cellular senescence: roles of the p53 and p16 pathways. EMBO J 2003;22:4212-22. [Crossref] [PubMed]
- van Deursen JM. Senolytic therapies for healthy longevity. Science 2019;364:636-7. [Crossref] [PubMed]
- Sieben CJ, Sturmlechner I, van de Sluis B, et al. Two-Step Senescence-Focused Cancer Therapies. Trends Cell Biol 2018;28:723-37. [Crossref] [PubMed]
- Lee S, Schmitt CA. The dynamic nature of senescence in cancer. Nat Cell Biol 2019;21:94-101. [Crossref] [PubMed]


