
USO1 expression is dysregulated in non-small cell lung cancer
Introduction
Lung cancer is the leading cause of death worldwide with more than 2.09 million new diagnoses each year, and only 18% of lung cancer patients surviving more than 5 years (1). Non-small cell lung cancer (NSCLC) accounts for approximately 85% of all lung cancers (2). NSCLCs share a common cellular origin; however, they represent a heterogeneous group of cancers with different clinical behaviours and prognoses. Although the diagnostic and therapeutic techniques including surgery, chemotherapy, radiotherapy as well as targeted therapies for NSCLC have made significant progress, the 5-year overall survival (OS) for NSCLC patients remain low (3). Furthermore, the majority of tumours are detected at an advanced stage, leading to unsuccessful targeted treatment. Therefore, identifying a reliable diagnostic biomarker that can detect early stage NSCLC as well as development of new targeted therapies remains crucial.
The Golgi apparatus is central to post translational modification and trafficking of proteins and lipids within the cell. It traffics a large part of the proteome and as a result impacts on mitosis, apoptosis and migration (4). endoplasmic reticulum (ER) to Golgi vesicular transport packs cargo into secretory vesicles and directs it to the plasma membrane as well as extracellularly. These proteins include receptors involved in downstream signalling pathways, channels, extracellular matrix components and signalling molecules. Therefore, transport form the ER-Golgi is an essential step for completion of cellular functions such as signal transduction pathways and cell-cell interactions. It is not surprising then, that dysregulation of this tightly controlled intracellular function can lead to cancer development. Multiple studies have implicated ER-Golgi trafficking in promoting tumour progression through alterations in the secretome (5) and that increased ER-Golgi trafficking could result in enhanced protein transport and tumour progression (6), and may also facilitate cancer cell invasion and metastasis (7). How the Golgi apparatus integrates signals to alter morphology and trafficking kinetics under physiological and pathological conditions is still poorly defined but could represent a new target for cancer treatment.
USO1 (also called p115/TAP) is a protein that tethers vesicles in the ER-Golgi complex playing a critical role in vesicular trafficking and transport during interphase (8). It plays a critical role in both mitosis and apoptosis and is essential for mitotic spindle function through interacting with γ-tubulin (9). USO1 has been shown to be involved in the tumourigenesis of multiple cancers including gastric cancer (10), colorectal cancer (11), breast cancer (12), hepatocellular carcinoma (13) and multiple myeloma (14). USO1 has been shown to have higher levels of expression in these cancers and its role in tumourigenesis contributes to cell proliferation and evasion of apoptosis. Furthermore, multiple studies have identified USO1 exerts its oncogenic role by interfering with the Erk signalling pathway (13-15).
Currently, to our knowledge there have been no studies regarding the expression of USO1 and its clinical implications in NSCLC. The aim of this study was to therefore identify if USO1 is altered in NSCLC and involved in tumourigenesis. We used tissue microarrays (TMAs) containing resected lung resections of 204 patients with NSCLC for investigation of the prognostic value of USO1 expression in NSCLC. We also used online gene expression databases to further investigate and validate our findings regarding the potential role of altered USO1expression in NSCLC.
Our results show USO1 is significantly increased in the early stages of lung adenocarcinoma (LUAD) and high levels are associated with a better overall survival, therefore, making it a potential diagnostic biomarker and as a potential target for new therapeutics in LUAD. We present the following article in accordance with the REMARK reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-230/rc).
Methods
Patients and clinical samples
We used primary tumour tissue samples from patients diagnosed with NSCLC pathological stage I–IV at St James Hospital, Dublin, Ireland from the period 1999–2007. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and approved by the St James’s Hospital & Tallaght University Hospital Joint Research Ethics Committee (No. 041018/8804), and individual consent for this retrospective analysis was waived.
A total of 204 patients with NSCLC were included in this study. Clinicopathological and histopathological data (including age, sex, smoking status, histology, TNM stage, surgical procedure, tumour grade, and primary site) were collected and the clinicopathological characteristics of our cohort of patients are presented in Table 1.
Table 1
| Clinicopathological parameters | High expression (H score ≥188) | Low expression (H score <188) | P value |
|---|---|---|---|
| Histology | 0.801 | ||
| LUSC | 44 | 64 | |
| LUAD | 46 | 36 | |
| Sex | 0.019 | ||
| Female | 45 | 34 | |
| Male | 55 | 70 | |
| Age, years | 0.82 | ||
| <65 | 41 | 51 | |
| ≥65 | 59 | 53 | |
| Node | 0.39 | ||
| Positive | 41 | 48 | |
| Negative | 59 | 56 | |
| Tumour size, cm | 0.016 | ||
| ≥5 | 33 | 49 | |
| <5 | 67 | 55 | |
| Grade | 0.795 | ||
| 1 | 7 | 9 | |
| 2 | 58 | 52 | |
| 3 | 35 | 43 | |
| Stage | 0.077 | ||
| I | 55 | 45 | |
| II | 19 | 30 | |
| III | 26 | 28 | |
| IV | 0 | 1 | |
| Smoker | 50 | 50 | 0.704 |
| Ex-smoker | 38 | 40 | |
| Never smoker | 12 | 14 |
NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
The median age of patients was 66 (range, 29–86 years) at time of diagnosis. The majority of patients were male (61%). Most patients were current smokers at time of diagnosis (n=100), 78 were ex-smokers and 26 were never smokers.
Histopathological examination of resected tumours revealed that LUSC was the dominant histological subtype (n=108), 82 cases were LUAD, 7 cases of pleomorphic carcinoma, 3 cases were large cell and 4 cases were classified as adenosquamous.
Postoperative staging demonstrated that 100 cases were stage I, 49 cases were stage 2, 54 cases were stage 3 and 1 case was stage 4. Forty-one cases had T1 disease, 123 cases had T2, 27 cases had T3, and 9 cases had T4 disease. A total of 115 had N0 disease, 51 had N1, and 38 had N2 disease.
All surgically resected tumour specimens and control specimens were fixed with 10% formalin fixed paraffin embedded (FFPE). The tumours were staged according to the Union for International Cancer Control Tumour-Node-Metastasis (TNM) Classification of Malignant Tumours 8th edition (16,17) and histologically subtyped and graded according to the World Health Organization guidelines (18,19).
Microarray construction
Following ethical approval, a tissue microarray (TMA) containing quadruplicate cores (0.6 mm) of the 204 NSCLC patients was generated using a Beecher Manual Tissue Arrayer (Model MTA-1) and 4 µm sections used for immunohistochemical analysis of USO1.
Immunohistochemistry detection methods
Immunohistochemistry was performed on 4 µm TMA sections. Slides were deparaffinised, rehydrated, washed and quenched according to standard protocol. ULTRA cell conditioning (ULTRA CC1 Roche Cat# 05279801001), pH9.1, was used for heat induced epitope retrieval (HIER). For USO1 antibody staining, slides were incubated with rabbit monoclonal primary antibody [p115 (H-11): Santa Cruz Biotechnology Cat# sc-48363, RRID: AB_628059)] diluted in PBS (1:50) for 64 min at ambient temperature and stained using the OptiViewTM DAB IHC detection kit (Roche Cat# 760-700, RRID: AB_2833075) and OptiViewTM amplification kit (Roche Cat# 06396518001) on a Ventana BenchMark XT processor.
Assessment of immunohistochemistry
The immunostaining of TMA slides was assessed by two pathologists without being aware of the clinical, pathological and follow-up data. Staining intensity was designated as either 0, 1+, 2+ or 3+ and each tumour section was given a H-score between 0–300 = 3(% at 3+) + 2(% at 2+) +1(% at 1+).
Tumours with high USO1 expression were designated as those with an average H score above the median value and low expression below the median. The median score obtained was 188.
In silico analysis of public gene expression datasets
Expression analysis and survival analysis
The Lung Cancer Explorer (LCE) database (20) was queried (https://lce.biohpc.swmed.edu/lungcancer/) using the gene symbol ‘USO1’ to conduct a meta-analysis of standardized mean difference (tumour vs. normal) in gene expression in NSCLC.
LCE was also used to provide the summary statistics for tumour-normal Gene expression differences for meta-analysis of standardized mean difference (tumour-normal) using Hedges’ G as an effect size metric. Summary statistics for survival and gene expression associations were based on univariate Cox Proportional-Hazards modelling. For all studies included for meta-analysis must have at least 10 samples in each group and meta-analysis was only performed for genes with data available from at least three qualifying studies. The analysis was performed separately for lung adenocarcinoma and squamous cell carcinoma.
Then, subgroup analysis of the mRNA expression of USO1 was conducted using the UALCAN database (http://ualcan.path.uab.edu) (21). The LUAD and lung squamous cell carcinoma (LUSC) datasets from The Cancer Genome Atlas (TCGA) were selected for analysis. USO1 mRNA expression was analysed in the following subgroups: sex, age, race, smoking status, cancer stage, tumour grade, and nodal status, metastasis, and TP53 mutation status. We then evaluated prognostic significance of USO1 in LUAD and LUSC using the Kaplan-Meier (KM) Plotter database (http://kmplot.com) (22).
Altered protein expression in the TCGA-LUAD was validated using UALCAN (21) to interrogate the CPTAC discovery dataset which investigated 111 tumours, (with 102 tumours paired with normal adjacent tissue samples) and subjected these samples to global proteome and phosphorproteome analysis (23).
Associations with immune infiltration analysis
For evaluation of any correlations between USO1 expression and immune cell infiltrates we utilized the TIMER (https://cistrome.shinyapps.io/timer/) (24) to plot purity-corrected partial Spearman’s rho value and statistical significance for the following immune cell infiltrates: B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils and dendritic cells.
Associations with mutated genes
To identify mutated genes from the associated TCGA datasets that result in changes of expression of USO1 in NSCLC an analysis was conducted using the MuTarget platform (https://www.mutarget.com/) with a mutation prevalence set at 2% (25), and validated using correlation analysis on TIMER2 portal (http://timer.cistrome.org/) (26).
Effects of copy number variations (CNV) on USO1
To examine whether the altered expression of USO1 was correlated with CNVs, the datasets for TCGA-LUAD and TCGA-LUSC PanCancer Atlas datasets (27) were analysed through cBioPortal (28) with the following parameters selected: mRNA expression z-scores relative to normal samples (log RNA Seq V2 RSEM), and Putative copy-number alterations from GISTIC. mRNA vs. CNA was plotted with Log2 copy number against mRNA expression z-scores relative to normal samples (log RNA Seq V2 RSEM).
Protein-protein interaction network analysis
Protein-protein interactions with USO1 were queried using the String database (http://string-db.org) (29) (interaction score >0.4). Correlation analysis of the associated proteins in normal tissue, LUAD and LUSC were subsequently analysed using LCE. The TCGA LUAD and LUSC databases were selected. All subsequent candidates were subsequently interrogated for differential gene expression in LUAD and LUSC using LCE and survival associations using KM plot.
Many more in silico analyses were run than can be included in the article, and their results are mentioned throughout the text. The interested reader can find them in a supplementary appendix online (Figures S1-S9).
All analyses were carried out using the default settings of the corresponding algorithms if not otherwise indicated, with the detailed dataset information and guidelines available at each algorithm portal.
Statistical analysis
Statistical analysis was performed using either the SPSS 25.0. statistical software package (SPSS Inc., Chicago, IL, USA), or Graphpad Prism 5.01 (Graphpad Software, San Diego, CA, USA). Correlations between USO1 expression and given categorised parameters were evaluated using the nonparametric Mann-Whitney U-test (for two categories) or Kruskal-Wallis test (for multiple categories). Kaplan-Meier curves were performed for survival curves, and statistical analysis was assessed using the log-rank test. Overall survival was defined as the time from the date of surgery to death. Patients who were still alive or lost to follow-up, were treated as censored data in the survival analysis. Univariate analysis of overall survival was performed using Kaplan-Meier method. Multivariate Cox proportional hazard regression analysis was carried using the methodology described in (30) to assess the prognostic significance of USO1 and other clinicopathological characteristics on survival.
The correlation of gene expression was evaluated by Spearman’s correlation. Overall, 95% confidence intervals (CIs) were used throughout the analysis. Statistical significance was defined as P<0.05.
Results
Pattern of USO1 expression in NSCLC and correlation to clinicopathological parameters
Positive immunohistochemical staining for USO1 showed nuclear, membranous and cytoplasmic staining pattern in NSCLC as shown in Figure 1.
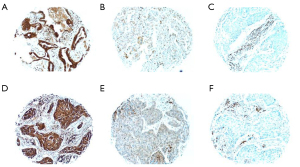
Background normal lung had weak granular cytoplasmic staining for USO1 Figure 2.
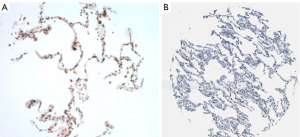
There was strong positive staining in background inflammatory cells including plasma cells and lymphocytes. Each of the four sections of tumour per case was given a H score and an average score of the four was calculated. After all cases were scored the median H score was calculated as 188. Tumours with scores ≥188 were designated as having high expression and those with scores <188 as having low expression of USO1.
The relationship between USO1expression and clinicopathological features was assessed and the results are presented in Table 1. Of the 204 cases 46/82 (56%) of LUAD and 44/108 (41%) of LUSC had expression above the median value (H score >188).
There was no significant correlation between USO1 expression and age, lymph node status, grade, stage, smoking status, or tumour cell type. Females and tumour size <5 cm were associated with higher USO1 expression compared to males and tumour size ≥5 cm.
The results of USO1 expression were analysed with regard to overall survival (OS) time of the patients and are presented in Figure 3.
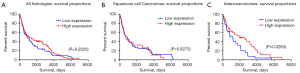
Relationship between USO1 expression and clinical outcome in NSCLC: univariate and multivariate survival analysis
At the time of analysis, the number of deaths that occurred was 164. Univariate survival analysis (log-rank test) for all histologies demonstrated significant association between OS and age ≥65, smoking status, tumour size ≥5 cm, stage, and nodal status (Table 2).
Table 2
| Parameter | Patients (cases) | Mean survival (months) | Median survival (months) | P value |
|---|---|---|---|---|
| USO1 expression (low vs. high) | 104 (low) | 53.6 | 29.9 | 0.202 |
| 100 (high) | 65.1 | 37.3 | ||
| Sex (male vs. female) | 125 (male) | 53.4 | 25.9 | 0.239 |
| 79 (female) | 65.9 | 45.0 | ||
| Age (<65 vs. ≥65) | 93 (<65) | 74.0 | 45.6 | 0.006 |
| 111 (≥65) | 48.9 | 25.4 | ||
| Histology (LUAD vs. LUSC vs. other) | 82 (LUAD) | 67.57 | 50.6 | 0.067 |
| 108 (LUSC) | 53.4 | 24.13 | ||
| 14 (other) | 39.3 | 60.1 | ||
| Smoking status (smoker vs. non-smoker) | 179 (smoker) | 61.6 | 35.7 | 0.011 |
| 25 (non-smoker) | 27.7 | 13.5 | ||
| Size (<5 vs. ≥5 cm) | 121 (<5 cm) | 71.9 | 84.9 | <0.001 |
| 83 (≥5 cm) | 42.3 | 16.0 | ||
| Stage (I, II, III, IV) | 100 (I) | 70.7 | 54.3 | <0.001 |
| 49 (II) | 57.2 | 27.1 | ||
| 54 (III) | 39.7 | 13.5 | ||
| 1 (IV) | 3.7 | 3.7 | ||
| Nodal status (positive vs. negative) | 89 (pos) | 48.4 | 18.6 | 0.025 |
| 115 (neg) | 66.3 | 48.9 |
NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Overall USO1 expression did not reach significance (P=0.2025) (Figure 3A). Univariate analysis in the LUAD group (n=82) demonstrated significant association between OS and the status of USO1 (P=0.0283) (Table 3).
Table 3
| Parameter | LUAD (n=82) | LUSC (n=108) |
|---|---|---|
| USO1 expression | 0.027 | 0.827 |
| Sex (male vs. female) | 0.122 | 0.826 |
| Age (≥65 vs. <65) | 0.087 | 0.102 |
| Smoking status (smoker vs. non-smoker) | 0.158 | 0.004 |
| Size (≥5 vs. <5 cm) | 0.122 | <0.001 |
| Stage (I, II, III, IV) | 0.021 | <0.001 |
| Nodal status (positive vs. negative) | 0.003 | 0.25 |
LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Patients with high level expression of USO1 had a better prognosis than those with low-level expression. Figure 3C shows a Kaplan-Meier survival curve in relation to USO1 expression in patients with LUAD. Nodal status and stage reached statistical significance also in the LUAD group (P=0.021 and P=0.003, respectively) (Table 3).
There was no significant difference in OS and USO1 expression when looking at the LUSC group (n=108) on univariate analysis (Figure 3B, Table 3) (P=0.8275).
To evaluate if USO1 protein expression is an independent prognostic factor in LUAD, a multivariate analysis using the Cox proportional hazard model was performed and included variables with P<0.05 in the univariate analysis, which included nodal status and stage. Multivariate analysis proved USO1 expression as an independent prognostic factor of overall survival in the LUAD group, with high expression associated with better OS (P=0.048, 95% CI: 0.351–0.995; Table 4).
Table 4
| Parameter | P value | Hazard ratio | 95% CI |
|---|---|---|---|
| USO1 expression (low vs. high) | 0.048 | 0.591 | 0.351–0.995 |
| Stage (I, II, III, IV) | 0.880 | 1.056 | 0.175–2.189 |
| Nodal status (positive vs. negative) | 0.456 | 0.619 | 0.520–2.146 |
Analyses were carried out using the methodology described in (30). CI, confidence interval; LUAD, lung adenocarcinoma.
Confirmation that USO1 has high mRNA and protein expression in LUAD and LUSC
A meta-analysis was performed using LCE to obtain an overview of tumour vs. non-malignant tissue (normal) differential gene expression of USO1 from multiple NSCLC datasets (20), and the results show that USO1 is significantly altered in LUAD (P.adj =3.3×10−10), while no significant altered expression of USO1 is observed for LUSC (P.adj =0.075) (Table S1).
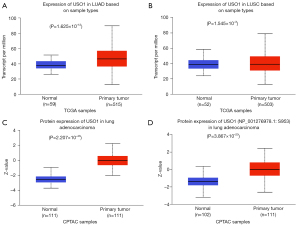
Further assessment of USO1 mRNA expression in LUAD and LUSC was done using UALCAN (using the TCGA dataset) confirming that USO1 mRNA is significantly elevated in both LUAD (Figure 4A) and LUSC (Figure 4B). We then looked at USO1 protein expression in LUAD on UALCAN using data from the Clinical Proteomic Tumour Analysis Consortium (CPTAC) dataset (31). We found that both total USO1 protein (Figure 4C) and phospho protein at position S953 (Figure 4D) are significantly upregulated in LUAD compared to normal. We further confirmed these observations using cProSite, and also demonstrated that total USO1 and phosphor s953 were also significantly elevated in LUSC (Figure S1). The elevated phosphorylation observed in the USO1 protein in LUAD and LUSC tumours is at position S953, found in the COOH-terminal acidic domain of USO1 and necessary for correct Golgi localization (32). As such this therefore suggests that the elevated levels of S953 phosphorylation may be associated with a functional role for USO1 in LUAD. Overall, the results from these different portals and from our TMA analysis all show that USO1 mRNA and protein expression is significantly increased in both LUAD and LUSC.
High levels of USO1 are associated with better overall survival in LUAD
Results from our TMA analysis showed that overall USO1 overexpression was not associated with better overall survival (Figure 3A). However, when stratified according to histology no OS benefit was observed for LUSC (Figure 3B), whereas a significant OS benefit was observed for patients with LUAD (Figure 3C). To further investigate this result we then used the online resource KM plotter (22) to evaluate the relationship between USO1 mRNA expression and OS and PFS in LUAD and LUSC.
In this larger dataset high expression of USO1 mRNA is associated with a better OS overall (Figure 5A). However, when stratified according to histology again this is restricted solely to the LUAD subgroup (n=719; HR, 0.47; 95% CI: 0.57–0.6, P<0.001; Figure 5B), whilst in the LUSC subgroup mRNA expression in USO1 had no impact on survival (n=524; HR, 1.08; 95% CI: 0.85–1.37, P=0.52, Figure 5C). These results mirror and validate the results on protein expression of USO1 in our TMA dataset as shown in Figure 3.
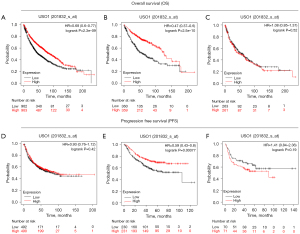
Similar results are seen when investigating progression free survival [defined as FP (first progression)] as shown in Figure 5. In this regard no FP benefit is observed overall (Figure 5D; HR, 0.93; 95% CI: 0.76–1.12, P=0.42), but in the LUAD subgroup (n=461) high USO1 mRNA expression is associated with longer FP (HR, 0.59; 95% CI: 0.43–0.8, P<0.001, Figure 5E) while USO1 expression in LUSC (n=141) has no significant impact on FP (Figure 5F, HR, 1.41; 95% CI: 0.84–2.36, P=0.19).
Relationship between USO1 mRNA levels and clinical pathological characteristics in the TCGA NSCLC datasets
Our TMA analysis found there was no significant correlation between USO1 protein expression and age, lymph node status, grade, stage, smoking status, or tumour cell type (Table 1). Females and tumour size <5 cm were associated with higher USO1 expression compared to males and tumour size ≥5 cm. To further assess these possibilities, a subgroup analysis on the TCGA-datasets was conducted using the UALCAN database. Results showed that USO1 mRNA was overexpressed in all stages (I–IV) in LUAD compared to normal, but there was no significant difference between each stage (Figure S2). In the TCGA-LUSC dataset, significant elevated expression was only observed for stage I and stage II compared to normal (Figure S2). USO1 was overexpressed in both males (n=238) (P<0.001) and females (n=276) (P<0.001) (Figure S3). USO1 was overexpressed in age groups 41–60 years (n=90) (P<0.001), 61–80 years (n=149) (P<0.001), 80–100 years (n=32) (P<0.001). USO1 overexpression was not significant in the age group 21–40 years (n=12) (P=0.12). USO1 upregulation was significant in patients who were Caucasian (n=387) (P<0.001) and African-American (n=51) (P=0.04) but not in Asians (n=8) (P=0.054) (Figure S3). USO1 is overexpressed in patients regardless of smoking status between non-smokers (n=75), smokers (n=118), and reformed smokers (n=303) all having significantly high levels of USO1 mRNA expression (Figure S3). There was no difference in USO1 mRNA levels between these subgroups. USO1 was overexpressed in patients with N0 (n=331), N1 (n=96) (metastasis in 1 to 3 axillary lymph nodes), N2 (n=74) (metastasis in 4 to 9 axillary lymph nodes), but not N3 (n=2) (metastasis in more than 10 axillary lymph nodes) lymph node metastasis (Figure S3). USO1 was significantly upregulated in both LUADs with (n=233) and without (n=279) TP53 mutations (P<0.001 in both groups). There was no significant difference in USO1 expression between these two groups (P=0.5) (Figure S3).
These results suggest that USO1 overexpression occurs as an early event in LUAD with significant upregulation occurring in stage 1 and N0 status. There is no significant difference in any two stages. USO1 upregulation occurs regardless of smoking status, sex and TP53 mutations status and in age groups >40 and in Caucasians and African-Americans. Similar results were seen in our TMA protein expression analysis of USO1.
Correlations between USO1 expression, copy number variation and mutations in NSCLC
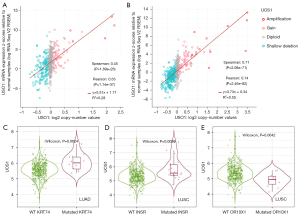
As USO1is significantly dysregulated in NSCLC, to further study the potential effects of this dysregulation we used cBioPortal (28) to assess for any correlations between copy number variations (CNVs) and gene expression changes in the TCGA-LUAD and -LUSC datasets. As shown in Figure 6, positive correlations between CNV and USO1 gene expression were observed both LUAD (Figure 6A) and LUSC (Figure 6B).
Next we assessed whether USO1 overexpression correlated with commonly mutated genes, or driver mutations in NSCLC. Using TIMER2 (26), the genes for p53, KRAS, ERBB2, EGFR, ALK, and PIK3CA were assessed to determine whether mutations within these key genes correlated with altered USO1 expression levels. The results showed that none of these genes when mutated correlate to altered USO1 overexpression in LUAD or LUSC (P>0.05). However, positive correlations between USO1 and wild-type KRAS, EGFR, ERBB2 and PIK3CA were observed for both LUAD and LUSC (Table 5).
Table 5
| Gene | LUAD | LUSC | ||||
|---|---|---|---|---|---|---|
| Partial cor. | P value | Partial cor. | P value | |||
| USO1 | TP53 | 0.0347 | 2.9×10−1 | 0.0370 | 0.673439 | |
| KRAS | 0.428 | 4.25×10−23 | 0.213 | 4.42×10−6 | ||
| EGFR | 0.368 | 8.46×10−17 | 0.260 | 1.44×10−8 | ||
| ERBB2 | 0.199 | 4.48×10−5 | 0.101 | 0.05 | ||
| PIK3CA | 0.520 | 4.46×10−35 | 0.353 | 2.56×10−15 | ||
| ALK | 0.073 | 0.205 | 0.087 | 0.14623117 | ||
Analysis was conducted using TIMER 2.0. Results are presented as purity-corrected partial Spearman’s rho value and statistical significance. Correlation cut-off values of the Spearman coefficient was set to R>0.33 (positive correlation) and R<-0.33 (negative correlation). Neg: negative correlation (P<0.05, P<0); Pos: Positive correlation (P<0.05, P>0); ns, not significant (P>0.05). NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Although there was no correlation seen for expression levels for mutated KRAS, EGFR, ERBB2 and PIK3CA in LUAD, overall expression for these genes is significantly correlated with USO1 mRNA levels. As USO1 is involved in vesicular transport, this may reflect a role in transporting tyrosine kinases or their interacting proteins important for tumourigenesis in LUAD to their final destination.
In addition to these frequently mutated genes, we further analysed whether mutations in any other genes may affect USO1 expression in LUAD and LUSC using muTarget (25). The results from muTARGET indicated that mutations in PLXND1 and KRT74 were associated with elevated USO1 expression in LUAD, while in LUSC mutations in INSR and SLIT1 correlate with elevated USO1 and mutated OR10X1 was found to be associated with decreased USO1 mRNA expression (Figure S4). However, when validating these observations using TIMER2 it was found that only KRT47 (Figure 6C), INSR (Figure 6D) and OR10X1 (Figure 6E) remained significant.
Correlations between USO1 expression and tumour mutational burden (TMB)
Given that TMB is widely associated as a potential biomarker for predicting the efficacy of immune checkpoint inhibitor therapy (33), we analysed the correlation between USO1 expression and various candidate markers of TMB as per (34). The results are presented in Table 6.
Table 6
| Variable | LUAD | LUSC | ||||
|---|---|---|---|---|---|---|
| R | P value | R | P value | |||
| DNA damage response (DDR) pathway | BRCA1 | 0.3 | 9×10−13*** | 0.14 | 0.0017** | |
| ATM | 0.52 | 2.8×10−38*** | 0.36 | 1.6×10−17*** | ||
| ATR | 0.23 | 3.9×10−8*** | 0.12 | 0.0067** | ||
| CDK1 | 0.14 | 0.0012** | 0.017 | 0.69 | ||
| CHEK1 | 0.24 | 2.2×10−8*** | 0.13 | 0.0018** | ||
| CHEK2 | 0.076 | 0.075 | 0.01 | 0.82 | ||
| TP53 | 0.19 | 9.9×10−6*** | 0.084 | 0.053 | ||
| Combined signature | 0.33 | 5.5×10−15*** | 0.15 | 0.00045*** | ||
| Mismatch excision repair (MMR) related genes | PMS2 | 0.5 | 1.6×10−35*** | 0.28 | 1.8×10−11*** | |
| MLH1 | 0.44 | 2.44×10−27*** | 0.33 | 4.1×10−15*** | ||
| MSH2 | 0.44 | 6.2×10−27*** | 0.21 | 9.3×10−7*** | ||
| MSH3 | 0.57 | 3.5×10−48*** | 0.41 | 5.41×10−23*** | ||
| MSH6 | 0.5 | 2.3×10−35*** | 0.25 | 4.3×10−9*** | ||
| PCNA | 0.17 | 5.4×10−5 *** | 0.016 | 0.7 | ||
| Combined signature | 0.57 | 1.3×10−48*** | 0.29 | 5.7×10−12*** | ||
Analysis was conducted using GEPIA2. Results are presented as Spearman’s rho value (R) alongside statistical significance. **, P<0.01; ***, P<0.001. LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Correlations between USO1 expression levels and the tumour microenvironment (TME) in LUAD and LUSC
The TME plays an important role in tumour growth, survival and ability to metastasize (35). The TME is composed of tumour cells, mixed immune cells and stromal cells communicating with each other to facilitate cancer progression. Components of the TME such as CD8+ and CD4+ tumour infiltrating lymphocytes (TILS) are associated with cancer prognosis. Moreover, the cell composition in the tumour immune microenvironment (TIME) of NSCLC significantly contributes to sensitivity of tumours to immune therapies (36,37).
We therefore sought to assess the correlation of USO1 expression in LUAD with different types of TILS, stromal cells, and markers of response to immune checkpoint inhibitors using TIMER (24). From this analysis we found that USO1 gene expression positively correlates with various immune infiltrates including CD8+ T cells, macrophages, neutrophils and dendritic cells (Table 7).
Table 7
| Variable | LUAD | LUSC | ||||
|---|---|---|---|---|---|---|
| Partial cor. | P value | Partial cor. | P value | |||
| USO1 | Purity | −0.00459 | 0.918922 | −0.07863 | 0.085915 | |
| B Cell | −0.01067 | 0.814866 | 0.102961 | 0.025293* | ||
| CD8+ T cell | 0.209888 | 2.98×10−6*** | 0.189294 | 3.29×10−5*** | ||
| CD4+ T cell | 0.056547 | 0.214305 | 0.095013 | 0.038453* | ||
| Macrophage | 0.16184 | 0.000345*** | 0.222949 | 8.72×10−7*** | ||
| Neutrophil | 0.218134 | 1.30×10−6*** | 0.253723 | 1.99×10−8*** | ||
| Dendritic cell | 0.171682 | 0.000138*** | 0.211603 | 3.36×10−6*** | ||
Analysis was conducted using TIMER. Results are presented as purity-corrected partial Spearman’s rho value and statistical significance. *, P<0.05; ***, P<0.001. Partial Cor. partial correlation (partial Spearman’s rho value). NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
However, when the effects of USO1 mRNA on immune infiltrates and associated survival were assessed, B cells and dendritic cells were the only TILs that were found to correlate with USO1 gene expression and patient survival (Table 8).
Table 8
| Variable | LUAD, P value | LUSC, P value | |
|---|---|---|---|
| USO1 | B cell | 0.000268*** | 0.778204 |
| CD8+ T cell | 0.345905 | 0.370702 | |
| CD4+ T cell | 0.507773 | 0.142871 | |
| Macrophage | 0.110109 | 0.651048 | |
| Neutrophil | 0.081069 | 0.126999 | |
| Dendritic cell | 0.047524* | 0.324067 | |
| USO1 | 0.765312 | 0.296457 |
Analysis was conducted using TIMER. Results are presented as purity-corrected partial Spearman’s rho value and statistical significance. *P<0.05; ***P<0.001. LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
USO1 associated protein-protein interactions have altered expression and prognostic value in NSCLC
Using STRING (29), we also identified a series of first neighbour protein-protein network for USO1 as presented in Figure S5. All of these proteins are associated with endoplasmic reticulum (ER) to Golgi transport, assembly and membrane stacking of the Golgi cisternae, forming inter-cisternal cross-bridges of the Golgi complex, and Golgi disassembly during mitosis (Figure S5). Using systematic meta-analysis in LCE we examined these genes for significant alterations in expression between tumour and normal lung, and the results are presented in Table S1.
Potential prognostic value of USO1 and associated genes in NSCLC
As upregulation of USO1 in LUAD is associated with better overall survival we next assessed if its first neighbours also had prognostic value. To assess this, we generated survival curves using KM plotter for patients with LUAD and LUSC (Figure S6 and Figure S7). In the LUAD subgroup better OS was associated with high mRNA expression levels for RAB1A; BET1; STXBP1L2 (SCFD1); GORASP1; GOS28 (GOSR1); and GOLGB1, whereas high mRNA expression was significantly associated with worse OS for STX5; YKT6; GOSR2 and GOLGA2. In contrast, altered mRNA expression in USO1s identified first neighbours had no impact on survival in the LUSC subgroup (P>0.05) as shown in Figure S6 and Figure S7.
These results suggest that dysregulation of genes involved in ER-Golgi transport plays a role in tumourigenesis in LUAD but not in LUSC.
Discussion
In this study we have shown that USO1 is overexpressed in both LUAD and LUSC, and that its upregulation is associated with better overall survival in LUAD but not LUSC. We have further linked that USO1 overexpression occurs in the early stages of LUAD and is independent of smoking status, sex, age >40, nodal status and stage. From in silico analyses we link USO1 and its first neighbours to have significant prognostic impact in LUAD. Finally, we find that USO1 upregulation in LUAD occurs independently of commonly mutated driver genes but is associated with various tumour immune infiltrates (particularly for CD8+ T Cells) and therefore could represent a new diagnostic biomarker for both prognosis and potentially stratify patients for checkpoint inhibitor therapy.
USO1 has been implicated in various tumour types including multiple myeloma (14), colorectal (11) and gastric cancer (10), but to our knowledge this is the first time that USO1 has been investigated in NSCLC. The mechanisms of how USO1 is important in tumourigenesis of LUAD are yet to be elucidated, and from our results further investigation is warranted.
Vesicular transport is vital for normal cellular function. Proteins are received from the ER and are transported to the Golgi where they undergo post translational modification, are processed and regulated before being directed to the appropriate target within a cell to control actions such as mitosis, apoptosis and migration. This is tightly regulated process, and it is not surprising that disruption of this system contributes to cancer progression, survival and metastasis.
Knockdown of USO1, which is critical for ER-Golgi transport in colorectal cancer and multiple myeloma cells inhibits cell proliferation and migration as well as inducing early apoptosis. Knockdown of USO1 also results in a decrease of cells in the G2-M phase of the cell cycle (11). Similarly, in gastric cancer overexpression of USO1 results in cell cycle arrest in the G0-G1 phase (10).
USO1 has been implicated in tumourigenesis in multiple cancers including gastric (10), colon (11), breast (12), liver cancer (13) as well as multiple myeloma (14) and leukaemia (38). Overexpression of USO1 has been shown to play a role in cell proliferation and cell cycle transition in these studies, and as such in general, one would expect high expression of USO1 to be associated with worse OS. However, survival analysis in tumours with USO1 overexpression has not generally been investigated. For example, in gastric cancer overexpression of USO1 promotes cell proliferation and G0-G1 to S phase transition however, overall survival was not assessed. Using KM-plotter, we have identified that high USO1 mRNA expression in gastric cancer is associated with a significantly better overall survival (P=5.6×10−7; Figure S8) similar to our observed results in LUAD. Our study used a H-scoring system which assessed both staining intensity and the proportion of tumour cells staining for USO1 assigning a score of 0–300 in each case. This gives a dynamic range to quantify USO1 abundance. Using this method, we were able to categorize LUAD patients into high and low protein expressers based on the median value. Our study found that USO1 is overexpressed in LUAD as well as LUSC, however, upregulation is associated with superior overall survival in LUAD and not LUSC. We also found many of USO1’s first neighbours were also prognostic in LUAD (Figure S6) but not in LUSC using KM plotter (Figure S7). As these proteins are all involved in Golgi transport system, it suggests that Golgi transport has a lesser role in LUSC tumourigenesis. Our findings and previously reported studies suggest that USO1, although involved in tumourigenesis, is most likely due to involve regulation of Erk (10,14,15), and is associated with a superior overall survival in LUAD and gastric cancer. The exact reason for this remains to be elucidated and warrants further investigation. It may be that a tumour specific transcript of USO1 is a key element as recently described for hepatocellular carcinoma, where a specific mRNA variant USO1-T (RefSeq NM_003715.4, transcript variant 2) is associated with worse prognostic outcomes and an aggressive phenotype in HCC (13).
In our analysis of the CPTAC dataset we found that levels of phosphorylation at position Serine 935 (S935) were significantly elevated in LUAD tumours. Originally a site for phosphorylation at position Serine 942 (S942) was identified as being critical for regulating USO1s association with the Golgi membrane (39), and when phosphorylated USO1 is found exclusively in the cytosol (39). Two isoforms of USO1 exist, and when mapped S942 on isoform 2 (NP_003706.2), equates to S935 on isoform 1 (NP_001276978.1) as shown in Figure S9. USO1 phosphorylation occurs primarily in interphase but not in mitotic cells (39), and is essential for post-mitotic reassembly of the Golgi apparatus (40). Reanalysis of existing proteomic datasets demonstrated that USO1 and phosphorylated S953 on USO1 are significantly elevated in both LUAD and LUSC (Figure S1), indicating that USO1 is functionally active in both histological subsets. However, previous ultrastructural studies on the ER and Golgi complex in adenocarcinomas and squamous cell carcinomas have identified clear differences between the LUAD and LUSC subtypes. Electron microscopy (EM) of LUADs is very heterogeneous and reflects the histological heterogeneity of LUADs. They can be composed of cells resembling those of embryological derivation of the lower respiratory tract, type II pneumocytes, or Clara cells (41). Many, however, are composed of cells rich in cytoplasmic organelles and include a very well-developed Golgi complex, rough endoplasm reticulum and several mitochondria (41-43), In contrast, on EM squamous cell carcinomas show abundant tonofibrils converging on desmosomes and extending into the intercellular bridges. Keratinization is marked with increased number of tonofibrils in a perinuclear arrangement (where the Golgi is usually located). They present a reduced rough endoplasmic reticulum (RER) with only a few ER tubules and abundant intermediate filaments, and the cytoplasm contains relatively few organelles (41-43). As such, the differences in the ER-Golgi may explain to some degree the differences in OS benefit observed between these two subtypes, but will require further evaluation.
As previously mentioned, previous studies of USO1 have shown it also regulates Erk activity. Upon activation, Erk phosphorylates GRASP65 which forms a complex with USO1 and results in unstacking of the Golgi cisternae, facilitating protein transport to the plasma membrane (13,44). A recent study in multiple myeloma has shown that cells deficient for USO1 have reduced proliferation and increased apoptosis via regulation of the Erk pathway (14). Support for this comes from an additional study in MDS which has shown similar results, in that USO1 plays an oncogenic role by inactivating Raf/ERK signalling (15).
The Raf/Mek/Erk pathway plays important roles in regulating fundamental biological processes such as proliferation, survival, and differentiation, and mutations and/or dysregulation leading to activation of this pathway commonly occur in a large fraction of human cancers, including NSCLC (45). As USO1 has been implicated in the regulation of this pathway in multiple cancers it is therefore possible that this is the mechanism of how USO1 exerts its oncogenic role in LUAD.
In our study multiple genes involved in ER-Golgi transport (USO1 and its first neighbours) also have altered expression and weakened correlation with each other in both LUAD and LUSC, however, these alterations only impacted on prognosis in LUAD. This was seen in both our TMA analysis (for USO1 expression) and the in silico analysis using KM plotter. This may suggest that ER-Golgi transport has a greater impact on tumourigenesis in LUAD compared to LUSC. Several of USO1s first neighbours such as BET1, GOSR1, and YKT6 have been shown to have differential expression in various cancers including NSCLC (46-48). Such studies reflect our in silico findings showing that expression of YKT6 in NSCLC tumour samples are associated with shorter DFS and OS (48). Conflicting data for RAB1A has emerged whereby immunohistochemistry found that high protein expression was associated with poorer OS (49), whereas high mRNA appears to be associated with better OS (Table S1).
Despite the advancements in lung cancer treatment, the survival rate among patients remains poor (3). This is, in part due to advanced lung cancer at the time of diagnosis and the lack of an effective biomarker for the identification of lung cancer. Therefore, identifying a reliable diagnostic biomarker that can detect early stage LUAD as well as development of new targeted therapies is crucial. Our study showed that USO1 overexpression in LUAD occurs independently of commonly mutated genes and therefore may represent a novel diagnostic biomarker. Another finding in this study is that mutations in certain genes such as KRT74 lead to both upregulation and downregulation of USO1, although the exact interactions that lead to altered USO1 expression in LUAD cells mutated for KRT74 is unknown.
Interestingly GOSR1 has been shown to influence the sensitivity of lung cancer cells to cisplatin (50), and as high mRNA expression of this gene is associated with better OS in LUAD (Table S1), this may therefore reflect a cohort of patients which may respond better to cisplatin based chemotherapy regimens. In this regard, high expression of USO1 results in reduced sensitivity to an all-trans retinoic acid (ATRA) derivative in myelodysplastic syndrome cells, while loss of USO1 enhances sensitivity (15). Given the potential for ATRA to target the cancer stem cell niche in lung cancer, whilst resensitizing cells to cisplatin (51), drugs that could potentially target USO1 may have potential utility in such regimens. In this regard using the DepMAp PRISM repurposing Primary Screen (52) Pipenzolate (P=3.56×10−5 linear regression) was identified as a candidate drug which could be repurposed to target USO1 (data not shown).
The Golgi complex may represent a novel target for LUADs. Moreover, a recent study disrupting the Golgi apparatus via inhibition of ADP ribosylation factor 1 (ARF-1), a critical component within the Golgi apparatus, in NSCLC cells with both secondary (T790M/del19) and tertiary (C797S/T790M/del19) acquired resistance to EGFR tyrosine kinase inhibitors (53). Disruption of the Golgi apparatus downregulated MET expression potentially involved in the acquired resistance to EGFR-TKIs via bypassing the EGFR pathway (53). As Erk is an element of EGFR signalling and has been identified as an important resistance mechanism for EGFR TKIs (54), targeting either USO1 specifically or disrupting the ER/Golgi network with drugs such as m-COPA (53) may warrant further investigation in EGFR mutated NSCLC.
Finally, our analysis also links high expression of USO1 with CD8+ T cell infiltrates in the tumour microenvironment (Table 7). As CD8+ T cells show potential as a marker for immune checkpoint inhibitor response in NSCLC (55), it may be possible to use USO1 expression combined with CD8+ T cell infiltration to stratify patients for immune checkpoint therapy.
Conclusions
Our findings provide the first evidence of USO1 overexpression in LUAD. Overexpression occurs in the early stages of tumour progression, and high levels predict better prognosis. Taken together, these findings suggest USO1 may be a potential diagnostic biomarker for patients with LUAD. These results suggest that the role of aberrant expression of members of the Golgi/ER require further evaluation in NSCLC.
Acknowledgments
Funding: This research was supported by a Pathological Society Trainee’s Small Grant—“Investigating the expression of USO1 in NSCLC”—awarded to Dr Anna Keogh.
Footnote
Reporting Checklist: The authors have completed the REMARK reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-230/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-230/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-230/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-230/coif). AMB reports an unremunerated leadership role as the current President of Lung Cancer Europe (LuCE), and an honorarium from Roche (Ireland) for presentation at educational events. SC declares that costs for registration/travel to educational conference have been covered by funding from Merck Sharp & Dohme, Bristol Myers Squibb and Pfizer. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and approved by the St James’s Hospital & Tallaght University Hospital Joint Research Ethics Committee (No. 041018/8804), and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bade BC, Dela Cruz CS. Lung Cancer 2020: Epidemiology, Etiology, and Prevention. Clin Chest Med 2020;41:1-24. [Crossref] [PubMed]
- Basumallik N, Agarwal M. Small Cell Lung Cancer. StatPearls. Treasure Island (FL): StatPearls Publishing Copyright © 2021, StatPearls Publishing LLC.; 2021.
- Simeone JC, Nordstrom BL, Patel K, et al. Treatment patterns and overall survival in metastatic non-small-cell lung cancer in a real-world, US setting. Future Oncol 2019;15:3491-502. [Crossref] [PubMed]
- Guizzunti G, Seemann J. Mitotic Golgi disassembly is required for bipolar spindle formation and mitotic progression. Proc Natl Acad Sci U S A 2016;113:E6590-9. [Crossref] [PubMed]
- Halberg N, Sengelaub CA, Navrazhina K, et al. PITPNC1 Recruits RAB1B to the Golgi Network to Drive Malignant Secretion. Cancer Cell 2016;29:339-53. [Crossref] [PubMed]
- Howley BV, Howe PH. Metastasis-associated upregulation of ER-Golgi trafficking kinetics: regulation of cancer progression via the Golgi apparatus. Oncoscience 2018;5:142-3. [Crossref] [PubMed]
- Bui S, Mejia I, Díaz B, et al. Adaptation of the Golgi Apparatus in Cancer Cell Invasion and Metastasis. Front Cell Dev Biol 2021;9:806482. [Crossref] [PubMed]
- Nakamura N, Lowe M, Levine TP, et al. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell 1997;89:445-55. [Crossref] [PubMed]
- Radulescu AE, Mukherjee S, Shields D. The Golgi protein p115 associates with gamma-tubulin and plays a role in Golgi structure and mitosis progression. J Biol Chem 2011;286:21915-26. [Crossref] [PubMed]
- Li XJ, Luo Y, Yi YF. P115 promotes growth of gastric cancer through interaction with macrophage migration inhibitory factor. World J Gastroenterol 2013;19:8619-29. [Crossref] [PubMed]
- Sui J, Li X, Xing J, et al. Lentivirus-mediated silencing of USO1 inhibits cell proliferation and migration of human colon cancer cells. Med Oncol 2015;32:218. [Crossref] [PubMed]
- Howley BV, Link LA, Grelet S, et al. A CREB3-regulated ER-Golgi trafficking signature promotes metastatic progression in breast cancer. Oncogene 2018;37:1308-25. [Crossref] [PubMed]
- Yoon S, Choi JH, Shah M, et al. USO1 isoforms differentially promote liver cancer progression by dysregulating the ER-Golgi network. Carcinogenesis 2021;42:1208-20. [Crossref] [PubMed]
- Jin Y, Dai Z. USO1 promotes tumor progression via activating Erk pathway in multiple myeloma cells. Biomed Pharmacother 2016;78:264-71. [Crossref] [PubMed]
- Li S, Deng G, Su J, et al. A novel all-trans retinoic acid derivative regulates cell cycle and differentiation of myelodysplastic syndrome cells by USO1. Eur J Pharmacol 2021;906:174199. [Crossref] [PubMed]
- Detterbeck FC, Chansky K, Groome P, et al. The IASLC Lung Cancer Staging Project: Methodology and Validation Used in the Development of Proposals for Revision of the Stage Classification of NSCLC in the Forthcoming (Eighth) Edition of the TNM Classification of Lung Cancer. J Thorac Oncol 2016;11:1433-46. [Crossref] [PubMed]
- Chansky K, Detterbeck FC, Nicholson AG, et al. The IASLC Lung Cancer Staging Project: External Validation of the Revision of the TNM Stage Groupings in the Eighth Edition of the TNM Classification of Lung Cancer. J Thorac Oncol 2017;12:1109-21.
- Travis WD, Brambilla E, Burke AP, et al. Introduction to The 2015 World Health Organization Classification of Tumors of the Lung, Pleura, Thymus, and Heart. J Thorac Oncol 2015;10:1240-2. [Crossref] [PubMed]
- Travis WD, Brambilla E, Nicholson AG, et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- Cai L, Lin S, Girard L, et al. LCE: an open web portal to explore gene expression and clinical associations in lung cancer. Oncogene 2019;38:2551-64. [Crossref] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649-58. [Crossref] [PubMed]
- Lánczky A, Győrffy B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J Med Internet Res 2021;23:e27633. [Crossref] [PubMed]
- Gillette MA, Satpathy S, Cao S, et al. Proteogenomic Characterization Reveals Therapeutic Vulnerabilities in Lung Adenocarcinoma. Cell 2020;182:200-225.e35. [Crossref] [PubMed]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res 2017;77:e108-10. [Crossref] [PubMed]
- Nagy Á, Győrffy B. muTarget: A platform linking gene expression changes and mutation status in solid tumors. Int J Cancer 2021;148:502-11. [Crossref] [PubMed]
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509-14. [Crossref] [PubMed]
- Campbell JD, Alexandrov A, Kim J, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 2016;48:607-16. [Crossref] [PubMed]
- Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401-4. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 2021;49:D605-12. [Crossref] [PubMed]
- Lee HJ, Choe G, Jheon S, et al. CD24, a novel cancer biomarker, predicting disease-free survival of non-small cell lung carcinomas: a retrospective study of prognostic factor analysis from the viewpoint of forthcoming (seventh) new TNM classification. J Thorac Oncol 2010;5:649-57. [Crossref] [PubMed]
- Chen F, Chandrashekar DS, Varambally S, et al. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat Commun 2019;10:5679. [Crossref] [PubMed]
- Nelson DS, Alvarez C, Gao YS, et al. The membrane transport factor TAP/p115 cycles between the Golgi and earlier secretory compartments and contains distinct domains required for its localization and function. J Cell Biol 1998;143:319-31. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Feng H, Shen W. ACAA1 Is a Predictive Factor of Survival and Is Correlated With T Cell Infiltration in Non-Small Cell Lung Cancer. Front Oncol 2020;10:564796. [Crossref] [PubMed]
- Genova C, Dellepiane C, Carrega P, et al. Therapeutic Implications of Tumor Microenvironment in Lung Cancer: Focus on Immune Checkpoint Blockade. Front Immunol 2022;12:799455. [Crossref] [PubMed]
- Passaro A, Brahmer J, Antonia S, et al. Managing Resistance to Immune Checkpoint Inhibitors in Lung Cancer: Treatment and Novel Strategies. J Clin Oncol 2022;40:598-610. [Crossref] [PubMed]
- Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 2018;24:541-50. [Crossref] [PubMed]
- Jaiswal AK, Truong H, Tran TM, et al. Focused CRISPR-Cas9 genetic screening reveals USO1 as a vulnerability in B-cell acute lymphoblastic leukemia. Sci Rep 2021;11:13158. [Crossref] [PubMed]
- Sohda M, Misumi Y, Yano A, et al. Phosphorylation of the vesicle docking protein p115 regulates its association with the Golgi membrane. J Biol Chem 1998;273:5385-8. [Crossref] [PubMed]
- Dirac-Svejstrup AB, Shorter J, Waters MG, et al. Phosphorylation of the vesicle-tethering protein p115 by a casein kinase II-like enzyme is required for Golgi reassembly from isolated mitotic fragments. J Cell Biol 2000;150:475-88. [Crossref] [PubMed]
- Corrin B, Nicholson AG. Pathology of the Lungs E-Book. 3rd Edition ed. Churchill Livingstone; 2011.
- Lupulescu A, Boyd CB. Lung cancer: a transmission and scanning electron microscopic study. Cancer 1972;29:1530-8. [Crossref] [PubMed]
- Obiditsch-Mayer I, Breitfellner G. Electron microscopy in cancer of the lung. Cancer 1968;21:945-51. [Crossref] [PubMed]
- Wei JH, Seemann J. Remodeling of the Golgi structure by ERK signaling. Commun Integr Biol 2009;2:35-6. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Pudova EA, Krasnov GS, Kobelyatskaya AA, et al. Gene Expression Changes and Associated Pathways Involved in the Progression of Prostate Cancer Advanced Stages. Front Genet 2020;11:613162. [Crossref] [PubMed]
- Lee SH, Yoo HJ, Rim DE, et al. Nuclear Expression of GS28 Protein: A Novel Biomarker that Predicts Prognosis in Colorectal Cancers. Int J Med Sci 2017;14:515-22. [Crossref] [PubMed]
- Ruiz-Martinez M, Navarro A, Marrades RM, et al. YKT6 expression, exosome release, and survival in non-small cell lung cancer. Oncotarget 2016;7:51515-24. [Crossref] [PubMed]
- Huang T, Chen B, Wang F, et al. Rab1A promotes IL-4R/JAK1/STAT6-dependent metastasis and determines JAK1 inhibitor sensitivity in non-small cell lung cancer. Cancer Lett 2021;523:182-94. [Crossref] [PubMed]
- Sun NK, Huang SL, Chien KY, et al. Golgi-SNARE GS28 potentiates cisplatin-induced apoptosis by forming GS28-MDM2-p53 complexes and by preventing the ubiquitination and degradation of p53. Biochem J 2012;444:303-14. [Crossref] [PubMed]
- MacDonagh L, Santiago RM, Gray SG, et al. Exploitation of the vitamin A/retinoic acid axis depletes ALDH1-positive cancer stem cells and re-sensitises resistant non-small cell lung cancer cells to cisplatin. Transl Oncol 2021;14:101025. [Crossref] [PubMed]
- Corsello SM, Nagari RT, Spangler RD, et al. Discovering the anti-cancer potential of non-oncology drugs by systematic viability profiling. Nat Cancer 2020;1:235-48. [Crossref] [PubMed]
- Ohashi Y, Okamura M, Katayama R, et al. Targeting the Golgi apparatus to overcome acquired resistance of non-small cell lung cancer cells to EGFR tyrosine kinase inhibitors. Oncotarget 2018;9:1641-55. [Crossref] [PubMed]
- Yu D, Zhao W, Vallega KA, et al. Managing Acquired Resistance to Third-Generation EGFR Tyrosine Kinase Inhibitors Through Co-Targeting MEK/ERK Signaling. Lung Cancer (Auckl) 2021;12:1-10. [Crossref] [PubMed]
- Shirasawa M, Yoshida T, Imabayashi T, et al. Baseline PD-L1 expression and tumour-infiltrated lymphocyte status predict the efficacy of durvalumab consolidation therapy after chemoradiotherapy in unresectable locally advanced patients with non-small-cell lung cancer. Eur J Cancer 2022;162:1-10. [Crossref] [PubMed]


