
Cannabinoid receptor 2 expression in early-stage non-small cell lung cancers identifies patients with good prognosis and longer survival
Introduction
Lung cancer is a leading cause of cancer-related death in men and women, with over 83% of all cases being non-small cell lung cancer (NSCLC). Despite the development of new therapies, the 5-year survival of NSCLC patients is only 23% (1). To improve prognosis, patients should be allocated to appropriate treatments, which requires robust prognostic parameters for their stratification. Unfortunately, NSCLC is a rather heterogeneous disease with variable progression, and established prognostic parameters such as disease stage, histological grade, performance status, and tumor genetics do not guarantee reliable outcome prediction. Therefore, several prognostic factors, including tissue biomarkers, have been proposed for both early stage and advanced NSCLC (2,3). However, although predictive biomarkers are used to stratify patients for targeted therapies (4,5), no biomarkers are routinely used for prognostic patient stratification in clinical practice. Identifying new reliable prognostic biomarkers could improve the accuracy of outcome prediction, facilitate the design of effective follow-up strategies for individual patients, and improve our understanding of NSCLC development.
Cannabinoid receptor 1 (CB1) and cannabinoid receptor 2 (CB2) were successfully cloned in the early 90s (6,7) and their functions were elucidated using natural components of Cannabis sativa (such as Δ9-tetrahydrocannabinol) and synthetic analogs (8). The endocannabinoids anandamide and 2-arachidonoylglycerol were also identified and their regulatory effects were studied (9-12).
CB1 and CB2 receptors belong to the G protein-coupled receptor family. They are present in many tissues and organs but unlike CB1, CB2 receptors are strongly expressed in immune cells, particularly in B lymphocytes, monocytes, and neutrophils (13). Moreover, their mRNA has been found in the spleen, tonsils, and pulmonary endothelial cells (14,15).
The expression of the CB1 and CB2 receptors has been detected in various types of cancer cells (16-20) and shown to affect cancer prognosis and disease outcome positively or negatively depending on the type of cancer (21-23). Several models suggest that agonistic stimulation of cannabinoid receptors reduces cancer cell proliferation (24,25). Moreover, the endogenous cannabinoid system has recently emerged as a potential therapeutic target (26,27).
Studies on NSCLC cell lines and murine models have shown that both endogenous and synthetic cannabinoids inhibit carcinogenesis by various mechanisms (28-32), and Milian et al. recently showed that CB1 and CB2 gene expression may be associated with longer survival in a mixed population of NSCLC patients (33). CB1 and CB2 are thus likely to be implicated in modulating NSCLC progression, affecting survival. Moreover, their expression may have prognostic value. To test this hypothesis, we analyzed CB1 and CB2 gene and protein expression in human NSCLC tissue, focusing on its effects on the clinical outcomes of patients following radical surgery. We present the following article in accordance with the STROBE reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-247/rc).
Methods
Patients and sample collection
One hundred NSCLC patients (stage IA–IIIA) undergoing radical surgery were prospectively enrolled and biobanked between August 2009 and April 2013. The gene and protein expression of CB1 and CB2 in tumor tissue were analyzed retrospectively. Tumor tissues were collected during surgery and stored in RNAlater RNA Stabilization Reagent (Qiagen, Hilden, Germany) at −80 ℃ and as formalin-fixed paraffin-embedded (FFPE) samples. One patient undergoing radical surgery was excluded due to a missing sample and another was excluded due to zero housekeeping gene amplification, indicating poor tissue quality. Thus, 98 NSCLC patients (67 men and 31 women, aged 29–82 years) were included in the statistical analysis (Table 1). Additional paired tumor and tumor-free lung tissue samples from 10 NSCLC patients were analyzed independently to compare their expression of the CB1 and CB2 genes. Patients were clinically managed according to relevant national and international guidelines (34). Specifically, stage IA–IIIA NSCLC patients underwent radical surgical resection based on the consensus of a multidisciplinary tumor board. Stage IB–IIIA NSCLC patients received platinum-based adjuvant chemotherapy, while patients with microscopically positive resection margins (R1) underwent adjuvant radiotherapy. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics review board of University Hospital Olomouc and the Faculty of Medicine and Dentistry (IRB number 172/08) and all participants signed an informed consent form before the study enrollment.
Table 1
| Characteristics | CB1 gene (qRT-PCR) | CB2 gene (qRT-PCR) | CB1 protein (IHC) | CB2 protein (IHC) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pos./total (%) | P value | Pos./total (%) | P value | Pos./total (%) | P value | Pos./total (%) | P value | ||||
| Gender | |||||||||||
| Female | 4/31 (12.9%) | 0.503 | 26/31 (83.9%) | 0.656 | 11/26 (42.3%) | 0.756 | 6/27 (22.2%) | 0.295 | |||
| Male | 14/67 (20.9%) | 52/67 (77.6%) | 16/45 (35.6%) | 17/46 (37.0%) | |||||||
| Stage | |||||||||||
| IA | 9/31 (29.0%) | 0.369 | 29/31 (93.5%) | 0.047 | 12/22 (54.5%) | 0.028 | 5/22 (22.7%) | 0.779 | |||
| IB | 2/24 (8.3%) | 20/24 (83.3%) | 8/15 (53.3%) | 6/18 (33.3%) | |||||||
| IIA | 3/16 (18.8%) | 12/16 (75.0%) | 1/11 (9.1%) | 3/10 (30.0%) | |||||||
| IIB | 3/16 (18.8%) | 10/16 (62.5%) | 5/14 (35.7%) | 5/14 (35.7%) | |||||||
| IIIA | 1/11 (9.1%) | 7/11 (63.6%) | 1/9 (11.1%) | 4/9 (44.4%) | |||||||
| Grade | |||||||||||
| 1 | 1/5 (20.0%) | 0.739 | 5/5 (100.0%) | 0.286 | 2/2 (100.0%) | 0.023 | 0/2 (0.0%) | 1 | |||
| 2 | 7/32 (21.9%) | 27/32 (84.4%) | 12/23 (52.2%) | 7/22 (31.8%) | |||||||
| 3 | 9/57 (15.8%) | 42/57 (73.7%) | 12/43 (27.9%) | 14/46 (30.4%) | |||||||
| Histology | |||||||||||
| Adenocarcinoma | 7/40 (17.5%) | 0.24 | 34/40 (85.0%) | 0.147 | 15/31 (48.4%) | 0.288 | 12/32 (37.5%) | 0.171 | |||
| Large-cell carcinoma | 3/7 (42.9%) | 7/7 (100.0%) | 2/7 (28.6%) | 0/7 (0.0%) | |||||||
| Squamous-cell carcinoma | 8/51 (15.7%) | 37/51 (72.5%) | 10/33 (30.3%) | 11/34 (32.4%) | |||||||
Data are presented as n/N (%). CB1, cannabinoid receptor 1; qRT-PCR, quantitative reverse-transcriptase polymerase chain reaction; pos., positive; CB2, cannabinoid receptor 2; IHC, immunohistochemistry.
RNA purification
Total RNA was extracted from 20–40 mg of tumor tissue fixed in RNAlater using the Trizol (Molecular Research Center, Cincinnati, USA)/chloroform (Sigma-Aldrich s.r.o, St. Louis, USA) extraction method and resuspended in diethylpyrocarbonate (DEPC)-treated water (Ambion, Austin, TX, USA) according to the manufacturer’s instructions. RNA concentration and purity were assessed using a Nanodrop ND 1000 instrument (ThermoScientific, Wilmington, DE, USA).
Reverse transcription
Reverse transcription was performed on 3 µg of total RNA using random primers (Promega, Madison, WI, USA), RNAsin ribonuclease inhibitor (Promega), and RevertAid H Minus M-MuLV Reverse Transcriptase (Fermentas, Vilnius, Lithuania) in a 30 µL reaction volume according to the manufacturer’s instructions. Samples were stored at −20 ℃ until quantitative polymerase chain reaction (qPCR) analysis.
Quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR)
qRT-PCR reactions were performed on LightCycler 1536 Multiwell plates (Roche, Basel, Switzerland). In each reaction, 23.5 ng of cDNA was mixed with LightCycler 1536 DNA Probes Master (Roche), and the appropriate TaqMan Gene Expression Assay (Applied Biosystems, Foster City, CA, USA; CB1: Hs01038532_m1, CB2: Hs00275635_m1, ACTB: Hs99999903_m1). The reaction mixtures and samples were pipetted using an Echo Liquid Handler (Roche). The volume of the reaction mixture was 882.5 nL, and the volume of each sample was 117.5 nL. Each sample was applied to the plate in four replicates. Plates were amplified using a LightCycler 1536 instrument. The temperature and amplification time were set according to the protocol supplied with the TaqMan Gene Expression Assays, and 50 amplification cycles were performed. ACTB (coding for actin β) was amplified as a reference gene. Fluorescence signals and cycle threshold (CT) values were evaluated using the LightCycler 1536 Software, ver. 1.1. ΔCT values were calculated by normalization to ACTB.
Immunohistochemistry (IHC)
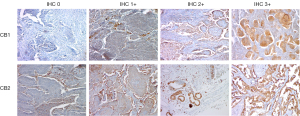
Since gene expression may not correlate with protein production, the presence of the CB1 and CB2 proteins in NSCLC tissue samples was validated immunohistochemically using a standardized 2 step protocol with diaminobenzidine as a chromogenic substrate. Formalin-fixed paraffin-embedded (FFPE) tissue samples from 82 patients in our cohort were of sufficient quality for analysis by IHC. Samples were stained using mouse monoclonal anti-CB1 antibody (ImmunoGenes, Cat# 01, RRID:AB_2910137) and mouse monoclonal anti-CB2 antibody (Abnova, Cat# H00001269-M01, RRID:AB_875479) according to the manufacturer’s protocol. Membranous and cytoplasmic staining was evaluated in at least three high power fields. Staining was categorized into four grades based on the proportion and intensity of positive tumor cells. H-scores were calculated using the following expression: [1× (percentage of grade 1+ cells) + 2× (percentage of grade 2+ cells) +3× (percentage of grade 3+ cells)], giving scores ranging from 0 to 300 (%). H-scores of 0 (0%) or 1 (<33%) indicate weak expression of CB1 and CB2, while H-scores of 2 (33–66%) or 3 (>66%) represent strong CB1 and CB2 expression. All immunostained samples were evaluated by an experienced pathologist blinded to the patients’ histological and clinical results.
Statistical analysis
Statistical analyses of CB1 and CB2 gene and protein expression using Pearson’s chi-square test, Fisher’s exact test, the logrank test, and stratified Cox regression were performed using R, ver. 3.5.0. Multivariate models of survival were generated in which age and gender were used as standard adjusting variables and the disease stage was used as a stratification variable. CB1 and CB2 expression were the independent variables of interest (Table S1). Additional models were generated in which body weight, body mass index (BMI) and chemotherapy were used as adjusting variables for survival (Table S2). The multivariate model for overall survival (OS) was non-convergent and thus could not be used. Specific cut-off values for CB1 and CB2 expression were determined using the maxstat function (maxstat R package, v. 0.7-25), which estimates cut-points based on the maximally selected log-rank statistic [using disease free survival (DFS) as an outcome variable] (35,36). Cut-off values corresponding to ΔCT 11.2 and 1.0 were used for CB1 and CB2 gene expression, respectively. The raw data are accessible at https://figshare.com (DOI 10.6084/m9.figshare.6321242).
Results
Survival analysis
NSCLC patients with tumors expressing the CB2 gene had significantly longer OS (log-rank test, P<0.001), cancer specific survival (CSS) [hazard ratio (HR) =0.3; 95% confidence interval (CI): 0.11–0.62; P=0.002], and DFS (HR =0.2; 95% CI: 0.11–0.48; P<0.001) than those whose tumors lacked CB2 expression (Figure 1A). However, survival was not affected by CB2 protein expression (Figure 1B), CB1 gene expression, or CB1 protein expression (Table S1).
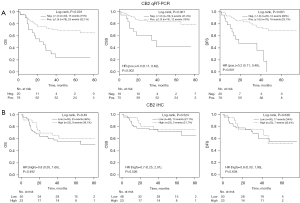
In a multivariate Cox model analysis stratified by disease stage, CB2 gene expression (but not CB2 protein expression) was identified as an independent prognostic factor for longer CSS (HR =0.274; P=0.013) and DFS (HR =0.322; P=0.009). Additionally, higher age was an independent prognostic factor for shorter CSS (Table S1). Including chemotherapy, weight, and BMI in the multivariate analysis did not affect these findings (Table S2, Figure S1). Univariate modelling indicated that the NSCLC histological subtype [adenocarcinoma (n=40), large-cell carcinoma (n=7) and squamous-cell carcinoma (n=51)] did not significantly influence survival (Figure S2A). Moreover, the expression of the CB1 and CB2 genes (measured by qRT-PCR) and the corresponding proteins (measured by IHC) was subtype-independent (Table 1). In the multivariate model for OS, the small number of large-cell carcinoma patients (n=7) resulted in a very broad HR CI, rendering this model’s output unreliable (data not shown). As expected, advanced disease stage was a negative prognostic factor for DFS, OS, and CSS (Figure S2B). In total, 40 (40.8%) of the 98 sampled NSCLC patients died over a median follow-up period of 44.5 months, 21 (22.3%) of them due to NSCLC; 63.2% of the patients survived for more than 3 years (Table S3).
CB1 and CB2 gene expression
CB2 gene expression was detected in 100% of the tumor tissue samples, of which 20.4% were classified as CB2-negative (ΔCT >1.0) and 79.6% as CB2-positive (ΔCT ≤1.0) based on the CB2 cut-off value. CB1 gene expression was detected in 50% of all tumor tissue samples, of which 63.3% were classified as CB1-negative (ΔCT >11.2) and 36.6% as CB1-positive (ΔCT ≤11.2) based on the CB1 cut-off value. We found higher CB2 gene expression in tumors of patients with a less advanced disease stage at the time of surgery (P=0.047). CB2 gene expression did not correlate with tumor histology or grading, while CB1 gene expression did not correlate with any clinical or morphological disease characteristics. Patients were categorized based on CB1 and CB2 gene and protein expression, gender, disease stage, tumor histology, and grading (Table 1). CB gene and protein expression in tumors was not found to be significantly related to body weight or BMI (Table 2). Moreover, neither body weight nor BMI affected survival in multivariate models (Table S2).
Table 2
| Characteristics | Weight (kg) | BMI (kg/m2) |
|---|---|---|
| CB1 gene (qRT-PCR) | ||
| Negative (>11.2) | 79 (68.75–92) | 27.6 (24.14–30.26) |
| Positive (≤11.2) | 70.5 (65–91) | 24.8 (22.85–30.96) |
| P value | 0.316 | 0.441 |
| CB2 gene (qRT-PCR) | ||
| Negative (>1.0) | 78 (67–92.75) | 26.3 (23.3–30) |
| Positive (≤1.0) | 78 (68–92) | 27.4 (23.87–31.48) |
| P value | 0.707 | 0.341 |
| CB1 protein (IHC) | ||
| Low | 78 (70–97) | 27.4 (24.33–30.5) |
| High | 81 (68–87.5) | 28.2 (23.61–31.76) |
| P value | 0.626 | 0.545 |
| CB2 protein (IHC) | ||
| Low | 78 (68–86.5) | 26 (23.69–30.37) |
| High | 81 (70–98.5) | 28.4 (25.67–32.18) |
| P value | 0.402 | 0.262 |
BMI, body mass index; kg, kilograms; CB, cannabinoid receptor; qRT-PCR, quantitative reverse-transcriptase polymerase chain reaction; IHC, immunohistochemistry.
To compare levels of cannabinoid receptors’ mRNA in tumor and normal tissues, we analyzed CB2 and CB1 gene expression in paired samples of tumor and tumor-free lung tissues in an independent cohort of 10 NSCLC patients, revealing that both the CB1 (P=0.008) and CB2 (P=0.056) genes were expressed at lower levels in tumor tissue samples than in tumor-free lung tissue (Figure 2).

CB1 and CB2 protein expression
The presence of CB1 and CB2 proteins was evaluated in FFPE tumor tissue samples (Figure 3) from 82 patients (stage I–IIIA) using IHC. High CB1 and CB2 protein expression (corresponding to IHC grades of 2 or 3) was present in 38% and 31.5% of samples, respectively. We found no significant differences in CB1 and CB2 protein expression in histological tumor subtypes (Table 1). In addition, non-tumor stromal and infiltrating cells exhibited very weak (IHC grade 0 or 1) CB1 protein expression and weak (IHC grade 1 or 2) CB2 protein expression (Figure 3). CB1 protein expression was associated with a less advanced disease stage (P=0.028).
CB1 and CB2 gene to protein correlation
Of the 71 patients analyzed for both CB1 gene and CB1 protein expression, 15 were qRT-PCR positive (21.1%) and 27 were IHC positive (38%). Of the 15 CB1 qRT-PCR positive patients, 8 were IHC negative (53.3%) and of the 56 CB1 qRT-PCR negative patients, 20 were IHC positive (35.7%) (P=0.634) (Table S4). Of the 73 patients analyzed for both CB2 gene and CB2 protein expression, 61 were qRT-PCR positive (83.6%) and 23 were IHC positive (31.5%). Of the 61 CB2 qRT-PCR positive patients, 44 were IHC negative (72.1%) and of the 12 CB2 qRT-PCR negative patients, 6 were IHC positive (50%) (P=0.176) (Table S4).
Discussion
We found that mRNA-level expression of CB2 but not CB1 is associated with significantly longer survival and fewer lymph node metastases at the time of surgery. Additionally, CB2 gene expression is a positive prognostic factor for CSS and DFS independently of age, gender, disease stage, tumor histology, and adjuvant chemotherapy treatment. Finally, tumors of patients with less advanced disease stage at the time of surgery showed higher CB2 gene expression. To our knowledge, this is the first study describing survival benefits of CB2 gene expression and its prognostic value in NSCLC patients undergoing radical surgery.
The only previously reported study on the effects of CB1 and CB2 gene expression on survival in NSCLC patients was conducted by Milian et al. (33). Unfortunately, several important factors render comparison of their results to ours difficult. First, our data were obtained only from patients undergoing radical surgery (up to stage IIIA), while Milian et al. also included patients with advanced (metastatic) disease. Moreover, Milian et al. did not report survival characteristics such as OS, DFS, CSS, the lengths of the follow-up and survival periods, the number of patients included in survival analysis, or the number of patients who died from NSCLC versus other causes. They found that both CB2 and CB1 gene expression improved survival, but controversially, they also concluded that disease stage did not affect survival. In contrast, we detected no effect of CB1 gene expression on survival. In fact, we observed no detectable CB1 expression in 50% of tumor samples and only weak expression in 63.3% of the remaining samples. Milian et al. correlated survival to mean CB1 and CB2 gene expression levels whereas we examined correlations between survival and weak or strong gene expression, using cut-off values determined by statistical analysis. We also correlated the expression of each gene to that of the corresponding protein (as determined by IHC). We therefore believe that our study provides more detailed and robust data on the influence of CB1 and CB2 gene and protein expression on clinical outcomes in NSCLC patients.
We detected CB1 and CB2 gene expression in 50% and 100% of the tumor tissue samples analyzed by qRT-PCR, respectively. Since the CB2 gene expression varied widely (from very weak to very strong), we divided our cohort into two groups based on a cut-off value for CB2 gene expression positivity; we assumed that patients with weak/very weak expression would have different survival characteristics to those with strong/very strong expression. Overall, 79.6% of our tumor samples were classified as CB2 expression-positive (i.e., having strong/very strong expression) and 20.4% as CB2 expression-negative (i.e., weak/very weak expression). We believe that this strategy enabled a more precise analysis because it reduced the likelihood that observed effects would be diluted due to the inclusion of patients with weak/very weak CB2 gene expression.
Interestingly, we observed no correlation of CB gene or protein expression with tumor histology, grading, or other factors reported to affect cannabinoid receptor expression (37,38). We also found that the positive effects of CB2 gene expression on OS, CSS and DFS in NSCLC are independent of other prognostic indicators. This suggests that its positive effects are related to the enhancement of CB2 gene transcription and/or mRNA stability by endogenous cannabinoids and/or other agonists because tumors with higher CB2 gene expression (and CB2 receptor production) are likely to be more responsive to such agents. This hypothesis is supported by several recent findings. First, Ravi et al. showed that combined treatment with the natural cannabinoid receptor agonist anandamide and an inhibitor of fatty acid amide hydrolase (FAAH; anandamide inactivating enzyme) reduces motility, migration and invasiveness of NSCLC cells. Combined treatment with anandamide and an FAAH inhibitor also induces G0/G1 cell cycle arrest, leading to apoptosis in NSCLC cells (32). Similar in vitro experiments showed that cannabinoids inhibit EGFR-induced AKT phosphorylation and induce apoptosis by up-regulating cyclooxygenase 2 (COX-2) and peroxisome proliferator-activated receptor gamma (PPAR-γ) (29,39). Moreover, Bremnes et al. reported that an increased presence of tumor-associated macrophages (TAM) or human lung-resident macrophages is associated with shorter survival (40), while Ravi et al. observed that TAM-induced epithelial-mesenchymal transition (EMT) and tumor growth is mitigated by the CB2 agonist JWH-015 in NSCLC (31). Finally, Milian et al. recently showed that the cannabinoid agonists tetrahydrocannabinol and cannabidiol inhibit proliferation, EMT, and migration in three types of lung cancer cells (A549, H460 and H1792) (33). It has also been reported that the effects of CB2 expression on survival vary widely in malignancies such as breast, skin, lymphoblastic, colon, hepatocellular and prostate cancers (17-20,24,30,41-46), suggesting that there are profound differences in the biology, signaling pathways, and immune cell interactions of different tumor types.
To better understand the relationship between tumorigenesis and CB1 and CB2 gene expression, we analyzed paired tumor and tumor-free tissue samples from an independent cohort of 10 NSCLC patients. Despite the small number of patients in this analysis, the tumor tissues clearly exhibited weaker expression of both CB1 and CB2 at the gene level (Figure 2). While reduced CB1 and CB2 gene expression could be a random consequence of tumorigenesis, we believe that it may represent an adaptive mechanism that allows NSCLC cells to minimize the inhibitory effects of endocannabinoids on their development.
Because IHC is widely used to measure protein expression in clinical practice, we also analyzed CB1 and CB2 protein expression by IHC. Both proteins were found to be expressed relatively weakly and their expression correlated poorly with that of the corresponding genes. This may be due to focal positivity and the significant heterogeneity of CB1 and CB2 protein expression across tumor sections. In addition, IHC is insufficiently sensitive to detect low but potentially biologically relevant concentrations of proteins, as demonstrated by the high frequency of IHC negativity in mRNA-positive tumors (47). The poor correlation between gene and protein expression may also be affected by post-translational mechanisms that modulate CB1 and CB2 protein activity and/or stability (48). Several classes of post-transcriptional regulators can affect protein expression, including small-noncoding RNAs, and four miRNAs targeting CB2 (hsa-mir-665, hsa-mir-3653-3p, hsa-mir-182-5p and hsa-mir-212-3p) were identified using the miRNet platform (https://www.mirnet.ca/Secure/MirTableView.xhtml). Moreover, Möhnle et al. experimentally verified that another miRNA, tsa-mir-665, significantly downregulates CB2 expression in human cardiomyocytes (49). We found no correlation between CB1 and CB2 protein expression and survival (Figure 1B), in accordance with Protein Atlas data (https://www.proteinatlas.org/ENSG00000188822-CNR2/tissue). However, in samples with detectable CB2 protein expression, it was mainly present in tumor but not stromal cells (Figure 3), suggesting that tumor cells are primary targets for cannabinoids. Given these findings and the increasing availability of molecular biology techniques such as qRT-PCR, we believe that measuring CB2 gene expression is a more accurate and clinically valuable way of monitoring cannabinoid receptor expression than protein-level analysis by IHC.
We recognize that our study has some limitations. First, we could not retrospectively analyze the history of recreational cannabis use in any of our patients, so we cannot exclude such use. However, cannabis was not commercially or medically available in the Czech Republic during our study, and recreational cannabis use is very rare among patients in the studied age group, so we assume the effect of this factor to be insignificant.
Second, endogenous cannabinoids may be over-produced in several types of cancers, which may increase their concentrations in tumor tissues and plasma (22), possibly affecting the course of the disease and the prognosis (27,50). Outcomes may also be affected by the activity of cannabinoid-metabolizing enzymes such as FAAH or γ-glutamyl hydrolase. Indeed, studies on prostate and breast cancer patients have shown that increased levels of hydrolytic enzymes are associated with poor prognosis (51,52). Moreover, two recent studies showed that an FAAH inhibitor had anti-invasive and antimetastatic effects in NSCLC cell lines (32,53). Therefore, the fact that we did not measure the concentrations of endogenous cannabinoids and the activity of cannabinoid-metabolizing enzymes is another potential limitation.
Despite these limitations, our results strongly indicate that CB2 gene expression is a useful prognostic parameter in NSCLC patients undergoing radical surgery. More studies on the modulation of CB2 gene expression and receptor activity are needed to elucidate their prognostic and therapeutic potential. Ideally, such studies should include measurements of endocannabinoid concentrations and the activity of cannabinoid-metabolizing enzymes.
Conclusions
In NSCLC patients undergoing radical surgery, mRNA-level expression of CB2 but not CB1 in tumor tissues is associated with significantly longer OS, CSS, and DFS as well as fewer lymph node metastases at the time of surgery. More studies are needed to evaluate the prognostic and therapeutic potential of CB2 expression as a biomarker of early-stage NSCLC.
Acknowledgments
Funding: This study was supported by Ministry of Health of the Czech Republic (No. NV18-03-00470); Ministry of Education, Youth and Sport of the Czech Republic (Nos. LM2018125, LM2018133); Palacky University Olomouc (No. LF 2021_019); Technological Agency of the Czech Republic (No. TN01000013); European Union - Next Generation EU (Programme EXCELES, ID Project No. LX22NPO5102) and Cancer Research Czech Republic.
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-247/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-247/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-247/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-247/coif). MV, SG, AR, JSro and MH report that they obtained institutional funding by Ministry of Health of the Czech Republic (No. NV18-03-00470), Ministry of Education, Youth and Sport of the Czech Republic (Nos. LM2018125, LM2018133), Palacky University Olomouc (No. LF 2021_019), Technological Agency of the Czech Republic (No. TN01000013) and European Union - Next Generation EU (Programme EXCELES, ID Project No. LX22NPO5102). JSro and MH are co-founders of spin-off company Intellmed, Ltd., and Cancer Research Czech Republic Foundation. EB, MK, PK report that they obtained institutional funding by Ministry of Health of the Czech Republic (No. NV18-03-00470), Ministry of Education, Youth and Sport of the Czech Republic (Nos. LM2018125, LM2018133) and European Union - Next Generation EU (Programme EXCELES, ID Project No. LX22NPO5102). PP, JC, MS, JK and JSka report that they obtained institutional funding by Ministry of Health of the Czech Republic (NV18-03-00470). The other author AP has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics review board of University Hospital Olomouc and the Faculty of Medicine and Dentistry (IRB number 172/08) and all participants signed an informed consent form before the study enrollment.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Miller KD, Nogueira L, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin 2019;69:363-85. [Crossref] [PubMed]
- Brundage MD, Davies D, Mackillop WJ. Prognostic factors in non-small cell lung cancer: a decade of progress. Chest 2002;122:1037-57. [Crossref] [PubMed]
- Chudacek J, Bohanes T, Klein J, et al. Detection of minimal residual disease in lung cancer. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub 2014;158:189-93. [Crossref] [PubMed]
- Gachechiladze M, Škarda J, Skanderová D, et al. Prognostic value of tumor-infiltrating lymphocytes (TILs) and their association with PD-L1 expression and DNA repair protein RAD51 in patients with resected non-small cell lung carcinoma. Lung Cancer 2020;147:30-8. [Crossref] [PubMed]
- Solomon B. Trials and Tribulations of EGFR and MET Inhibitor Combination Therapy in NSCLC. J Thorac Oncol 2017;12:9-11. [Crossref] [PubMed]
- Matsuda LA, Lolait SJ, Brownstein MJ, et al. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature 1990;346:561-4. [Crossref] [PubMed]
- Munro S, Thomas KL, Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature 1993;365:61-5. [Crossref] [PubMed]
- Pacher P, Bátkai S, Kunos G. The endocannabinoid system as an emerging target of pharmacotherapy. Pharmacol Rev 2006;58:389-462. [Crossref] [PubMed]
- Devane WA, Hanus L, Breuer A, et al. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science 1992;258:1946-9. [Crossref] [PubMed]
- Mechoulam R, Ben-Shabat S, Hanus L, et al. Identification of an endogenous 2-monoglyceride, present in canine gut, that binds to cannabinoid receptors. Biochem Pharmacol 1995;50:83-90. [Crossref] [PubMed]
- Pertwee RG. Emerging strategies for exploiting cannabinoid receptor agonists as medicines. Br J Pharmacol 2009;156:397-411. [Crossref] [PubMed]
- Sugiura T, Kondo S, Sukagawa A, et al. 2-Arachidonoylglycerol: a possible endogenous cannabinoid receptor ligand in brain. Biochem Biophys Res Commun 1995;215:89-97. [Crossref] [PubMed]
- Lu HC, Mackie K. An Introduction to the Endogenous Cannabinoid System. Biol Psychiatry 2016;79:516-25. [Crossref] [PubMed]
- Galiègue S, Mary S, Marchand J, et al. Expression of central and peripheral cannabinoid receptors in human immune tissues and leukocyte subpopulations. Eur J Biochem 1995;232:54-61. [Crossref] [PubMed]
- Atwood BK, Mackie K. CB2: a cannabinoid receptor with an identity crisis. Br J Pharmacol 2010;160:467-79. [Crossref] [PubMed]
- De Jesús ML, Hostalot C, Garibi JM, et al. Opposite changes in cannabinoid CB1 and CB2 receptor expression in human gliomas. Neurochem Int 2010;56:829-33. [Crossref] [PubMed]
- Caffarel MM, Andradas C, Mira E, et al. Cannabinoids reduce ErbB2-driven breast cancer progression through Akt inhibition. Mol Cancer 2010;9:196. [Crossref] [PubMed]
- Pérez-Gómez E, Andradas C, Blasco-Benito S, et al. Role of cannabinoid receptor CB2 in HER2 pro-oncogenic signaling in breast cancer. J Natl Cancer Inst 2015;107:djv077. [Crossref] [PubMed]
- Martínez-Martínez E, Gómez I, Martín P, et al. Cannabinoids receptor type 2, CB2, expression correlates with human colon cancer progression and predicts patient survival. Oncoscience 2015;2:131-41. [Crossref] [PubMed]
- Xu X, Liu Y, Huang S, et al. Overexpression of cannabinoid receptors CB1 and CB2 correlates with improved prognosis of patients with hepatocellular carcinoma. Cancer Genet Cytogenet 2006;171:31-8. [Crossref] [PubMed]
- Fraguas-Sánchez AI, Martín-Sabroso C, Torres-Suárez AI. Insights into the effects of the endocannabinoid system in cancer: a review. Br J Pharmacol 2018;175:2566-80. [Crossref] [PubMed]
- Hermanson DJ, Marnett LJ. Cannabinoids, endocannabinoids, and cancer. Cancer Metastasis Rev 2011;30:599-612. [Crossref] [PubMed]
- Ladin DA, Soliman E, Griffin L, et al. Preclinical and Clinical Assessment of Cannabinoids as Anti-Cancer Agents. Front Pharmacol 2016;7:361. [Crossref] [PubMed]
- Velasco G, Sánchez C, Guzmán M. Towards the use of cannabinoids as antitumour agents. Nat Rev Cancer 2012;12:436-44. [Crossref] [PubMed]
- Boyacıoğlu Ö, Bilgiç E, Varan C, et al. ACPA decreases non-small cell lung cancer line growth through Akt/PI3K and JNK pathways in vitro. Cell Death Dis 2021;12:56. [Crossref] [PubMed]
- Dhopeshwarkar A, Mackie K. CB2 Cannabinoid receptors as a therapeutic target-what does the future hold? Mol Pharmacol 2014;86:430-7. [Crossref] [PubMed]
- Velasco G, Hernández-Tiedra S, Dávila D, et al. The use of cannabinoids as anticancer agents. Prog Neuropsychopharmacol Biol Psychiatry 2016;64:259-66. [Crossref] [PubMed]
- Vidinský B, Gál P, Pilátová M, et al. Anti-proliferative and anti-angiogenic effects of CB2R agonist (JWH-133) in non-small lung cancer cells (A549) and human umbilical vein endothelial cells: an in vitro investigation. Folia Biol (Praha) 2012;58:75-80. [PubMed]
- Preet A, Ganju RK, Groopman JE. Delta9-Tetrahydrocannabinol inhibits epithelial growth factor-induced lung cancer cell migration in vitro as well as its growth and metastasis in vivo. Oncogene 2008;27:339-46. [Crossref] [PubMed]
- Preet A, Qamri Z, Nasser MW, et al. Cannabinoid receptors, CB1 and CB2, as novel targets for inhibition of non-small cell lung cancer growth and metastasis. Cancer Prev Res (Phila) 2011;4:65-75. [Crossref] [PubMed]
- Ravi J, Elbaz M, Wani NA, et al. Cannabinoid receptor-2 agonist inhibits macrophage induced EMT in non-small cell lung cancer by downregulation of EGFR pathway. Mol Carcinog 2016;55:2063-76. [Crossref] [PubMed]
- Ravi J, Sneh A, Shilo K, et al. FAAH inhibition enhances anandamide mediated anti-tumorigenic effects in non-small cell lung cancer by downregulating the EGF/EGFR pathway. Oncotarget 2014;5:2475-86. [Crossref] [PubMed]
- Milian L, Mata M, Alcacer J, et al. Cannabinoid receptor expression in non-small cell lung cancer. Effectiveness of tetrahydrocannabinol and cannabidiol inhibiting cell proliferation and epithelial-mesenchymal transition in vitro. PloS One 2020;15:e0228909. [Crossref] [PubMed]
- Ettinger DS, Akerley W, Bepler G, et al. Non-small cell lung cancer. J Natl Compr Canc Netw 2010;8:740-801. [Crossref] [PubMed]
- Lausen B, Schumacher M. Maximally Selected Rank Statistics. Biometrics 1992;48:73-85. [Crossref]
- Hothorn T, Lausen B. On the exact distribution of maximally selected rank statistics. Computational Statistics & Data Analysis 2003;43:121-37. [Crossref]
- Rossi F, Punzo F, Umano GR, et al. Role of Cannabinoids in Obesity. Int J Mol Sci 2018;19:2690. [Crossref] [PubMed]
- Verty AN, Stefanidis A, McAinch AJ, et al. Anti-Obesity Effect of the CB2 Receptor Agonist JWH-015 in Diet-Induced Obese Mice. PloS One 2015;10:e0140592. [Crossref] [PubMed]
- Ramer R, Heinemann K, Merkord J, et al. COX-2 and PPAR-γ confer cannabidiol-induced apoptosis of human lung cancer cells. Mol Cancer Ther 2013;12:69-82. [Crossref] [PubMed]
- Bremnes RM, Al-Shibli K, Donnem T, et al. The role of tumor-infiltrating immune cells and chronic inflammation at the tumor site on cancer development, progression, and prognosis: emphasis on non-small cell lung cancer. J Thorac Oncol 2011;6:824-33. [Crossref] [PubMed]
- Qamri Z, Preet A, Nasser MW, et al. Synthetic cannabinoid receptor agonists inhibit tumor growth and metastasis of breast cancer. Mol Cancer Ther 2009;8:3117-29. [Crossref] [PubMed]
- Casanova ML, Blázquez C, Martínez-Palacio J, et al. Inhibition of skin tumor growth and angiogenesis in vivo by activation of cannabinoid receptors. J Clin Invest 2003;111:43-50. [Crossref] [PubMed]
- McKallip RJ, Lombard C, Fisher M, et al. Targeting CB2 cannabinoid receptors as a novel therapy to treat malignant lymphoblastic disease. Blood 2002;100:627-34. [Crossref] [PubMed]
- Cianchi F, Papucci L, Schiavone N, et al. Cannabinoid receptor activation induces apoptosis through tumor necrosis factor alpha-mediated ceramide de novo synthesis in colon cancer cells. Clin Cancer Res 2008;14:7691-700. [Crossref] [PubMed]
- Adinolfi B, Romanini A, Vanni A, et al. Anticancer activity of anandamide in human cutaneous melanoma cells. Eur J Pharmacol 2013;718:154-9. [Crossref] [PubMed]
- Orellana-Serradell O, Poblete CE, Sanchez C, et al. Proapoptotic effect of endocannabinoids in prostate cancer cells. Oncol Rep 2015;33:1599-608. [Crossref] [PubMed]
- Wallander ML, Geiersbach KB, Tripp SR, et al. Comparison of reverse transcription-polymerase chain reaction, immunohistochemistry, and fluorescence in situ hybridization methodologies for detection of echinoderm microtubule-associated proteinlike 4-anaplastic lymphoma kinase fusion-positive non-small cell lung carcinoma: implications for optimal clinical testing. Arch Pathol Lab Med 2012;136:796-803. [Crossref] [PubMed]
- Börner C, Martella E, Höllt V, et al. Regulation of opioid and cannabinoid receptor genes in human neuroblastoma and T cells by the epigenetic modifiers trichostatin A and 5-aza-2’-deoxycytidine. Neuroimmunomodulation 2012;19:180-6. [Crossref] [PubMed]
- Möhnle P, Schütz SV, Schmidt M, et al. MicroRNA-665 is involved in the regulation of the expression of the cardioprotective cannabinoid receptor CB2 in patients with severe heart failure. Biochem Biophys Res Commun 2014;451:516-21. [Crossref] [PubMed]
- Tegeder I. Endocannabinoids as Guardians of Metastasis. Int J Mol Sci 2016;17:230. [Crossref] [PubMed]
- Thors L, Bergh A, Persson E, et al. Fatty acid amide hydrolase in prostate cancer: association with disease severity and outcome, CB1 receptor expression and regulation by IL-4. PloS One 2010;5:e12275. [Crossref] [PubMed]
- Shubbar E, Helou K, Kovács A, et al. High levels of γ-glutamyl hydrolase (GGH) are associated with poor prognosis and unfavorable clinical outcomes in invasive breast cancer. BMC Cancer 2013;13:47. [Crossref] [PubMed]
- Winkler K, Ramer R, Dithmer S, et al. Fatty acid amide hydrolase inhibitors confer anti-invasive and antimetastatic effects on lung cancer cells. Oncotarget 2016;7:15047-64. [Crossref] [PubMed]


