
Onco-biome in pharmacotherapy for lung cancer: a narrative review
Introduction
More than 100 trillion bacteria coexist in the gastrointestinal tract, and the flora contains 100 times as many genes as human genes (1,2). With the recent development of technology for metagenomic analysis and the refinement of sterile experimental animals, the composition of the flora and its role in immunity by cross-talk have been elucidated. Recently, accumulating scientific evidence has shown that dysbiosis in the intestinal flora causes a variety of diseases, including inflammatory bowel disease, diabetes, liver cirrhosis, and reflux esophagitis (3). In addition, the microbiome, comprising the bacteria, archaea, fungi, and viruses that cohabitate throughout the body, has been increasingly recognized for its important roles in multiple steps of cancer development or treatment in patients with cancer over the last five years. The “Hallmarks of Cancer” developed in 2022 details the crucial factors that should be the focus of future cancer research. In this review article, the discussion of the polymorphic microbiome that modulates the immune system in cancer patients expands the information detailed in “Hallmarks of Cancer” (4,5).
Lung cancer is the leading cause of cancer death worldwide, with 75,000 deaths annually, and half of the patients die within 1 year of diagnosis. Additionally, the 5-year survival rate in metastatic patients with lung cancer is less than 6% (6,7). In metastatic lung cancer patients, including small cell lung cancer (SCLC) and non-small-cell lung cancer (NSCLC) patients, the crucial treatment axis is pharmacotherapy, such as cytotoxic chemotherapy, molecular targeted agents (MTAs), and immunotherapy. These therapies can be used as single agents or in combinations of each drug modality (8).
The most important agents of pharmacotherapy currently used are MTA and immune checkpoint inhibitors (ICIs). MTA has specific (or limited) efficacy for kinase inhibitors in patients with specific genetic alterations, including those in EGFR, ALK rearrangement, ROS1 rearrangement, BRAF, RET rearrangement, KRAS G12C, and MET exon Δ14 skipping mutations (9-16), however, they are less effective with ICIs for those who do not have specific genetic alterations (17).
For SCLC, chemotherapy has long been the only treatment strategy, and for NSCLC, targeted therapies such as MTAs with gene mutations have been used. Therefore, the advent of ICIs has had a profound impact on lung cancer treatment. ICIs are characterized by the so-called “tail effect”, which is the long-term nature of the response observed once the patients do respond. The definitive biomarkers are programmed death-ligand 1 (PD-L1) expression on tumor cells or tumor mutation burden (18), but they are not sufficient. In addition, the toxicities are characterized as immune-related adverse events (irAEs), which are autoimmune-like reactions that are not characteristics of cytotoxic chemotherapy or MTA (19,20). Currently, studies of the gut microbiome are gradually revealing its role in cancer immunity by the collective effects of all the microorganisms that reside in all surfaces of the human body. Optimizing the microbiota composition leads to the modulation of immunity to improve the treatment efficacy of immunotherapy as a therapeutic intervention.
In this narrative review, we focus on the current status of knowledge of the onco-microbiome by the exploration of research from bench to bedside, studies of microbiome-based lung cancer treatment, and future perspectives for clinical applications.
Methods
To obtain relevant literature, we searched Medline (via PubMed) using the keywords “cancer”, and “microbiome” from the inception of the database to March 2022. The search summary is provided in Table 1. Our search was restricted to publications in the English language. We retrieved other eligible studies by manual searching of the reference list of included studies. For a search of current trials, a search was performed at Clinical Trials.gov (https://clinicaltrials.gov/). We present the following article in accordance with the Narrative Review reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-299/rc).
Table 1
| Items | Specification |
|---|---|
| Date of search | 1, March 2022 |
| Databases and other sources searched | PubMed |
| Search terms used | Cancer, microbiome |
| Timeframe | 1979–2022 |
| Inclusion and exclusion criteria | Inclusion criteria: English |
| Selection process | Akiko T. Tateishi and Yusuke Okuma independently conducted the selection; a consensus was obtained |
Microbiota and cancer development
Gut microbiota (GM)
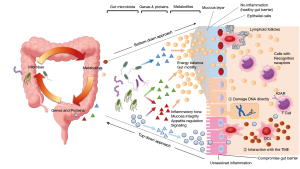
The GM is known to have a close relationship with human health and disease (1). The GM and the intestine are in an exquisite balance via genes, proteins, and metabolites of both bacteria and humans. Some bacteria improve energy balance and intestinal motility, maintain healthy intestinal mucosa, and improve the immune environment through metabolites, while others directly induce DNA destruction, intestinal mucosal disruption, and inflammatory triggering by the immune system through metabolites (Figure 1). In the last decade, with the development of next-generation sequencing and bioinformatic technologies, the relationship between microorganisms and cancer has been studied in more detail, and it has become clear that microorganisms are involved in protective or deleterious effects on cancer development and malignant progression (21). Certain microorganisms have been found to be involved in the carcinogenesis of colorectal cancer (22), liver cancer, biliary tract cancer (23), breast cancer (24,25), and stomach cancer (26). The mechanism of carcinogenesis is thought to be due to two effects of the microbiome. The first effect is mutagenesis of the colonic epithelium. This occurs with the production of bacterial toxins and other molecules that either damage DNA directly, disrupt the systems that maintain genomic integrity, or stress cells in other ways that indirectly impair the fidelity of DNA replication and repair (27). For example, E. coli carries the pks locus, which demonstrably mutagenizes the human genome and is implicated in conveying hallmark-enabling mutations (28). Secretion of butyrate acid, a metabolite of Porphyromonas sp., has been shown to contribute to tumorigenesis by inducing senescence of fibroblasts and epithelial cells (29). In addition to the aforementioned cancer types, the GM is thought to have an effect on other cancers, such as hematologic tumors or lung cancer, through interactions with the tumor microenvironment (TME) (30). Microbial mechanisms can modulate the activity of primary and secondary lymphoid organs of the gut epithelial barrier and regulate the immune tone of the TME (Figure 1).
A recent multicenter, multinational clinical trial demonstrated that higher diversity of intestinal microbiota is significantly associated with lower patient mortality after allogeneic hematopoietic stem cell transplantation (31). Moreover, analysis of more than 10,000 longitudinal fecal samples in an allogeneic hematopoietic stem cell transplantation trial revealed that immune reconstitution dynamics were closely related to GM composition (32). Links between GM, nutrition, posttransplant bone marrow (BM) and thymic cellularity, and lympho- and myelopoiesis have also been demonstrated in mouse models (33).
Various facts about the impact of the GM on adaptive immunity have also been elucidated. The intestinal ecosystem can influence both local and distant neoplasia by affecting the immune context, the influx of myeloid and lymphoid cells, and the patterns of inflammatory and metabolic processes. Secretory components of the GM can be important; for example, outer membrane vesicles (OMVs) can reprogram the TME toward a pro-TH1 phenotype (CXCL10, IFN-g) (34). Similarly, commensal microbiota was shown to prime tumor-associated innate myeloid cells for tumor necrosis factor-α (TNF-α) (IL-1b, IL-12, and Cxcl10) production in response to anti-IL-10R/CpG-ODN treatment, and antibiotics, germ-free, or TLR4-/- status attenuated this response and TNF-dependent early tumor necrosis. Anti-CTLA-4-induced gut barrier dysfunction was also found to be critical for the systemic translocation of Bifidobacterium-derived inosine, in turn promoting TH1 activation and antitumor immunity by agonizing T-cell-specific adenosine 2A receptor (A2AR) signaling in the context of dendritic cell (DC) costimulation (35). These examples demonstrate that barrier injury is accompanied by a deviation in the local microbiome or translocation of microbial metabolites, resulting in mobilization of DCs to gut-associated lymphoid tissues and contributing to infiltration of the tumor bed by activated helper or cytotoxic T cells. The TME is composed of a dense network of adrenergic nerve fibers that influence oncogenesis of brain and non-brain tumors as well as immune components derived from stromal, tumor, endothelial cells, and hematopoietic progenitor–derived immune component (36,37). Enteric nervous system neurons are both affected by the GM and functionally tuned according to their location in the gut. A subset of microbiota-responsive neurons was found to influence metabolic control independent of the central nervous system (38). These findings suggest close relationships between mucosal or tumoral commensals and tumor innervation, which warrants further study.
Lung microbiota
Lung cancer is closely associated with chronic inflammation, but the causes of inflammation and specific immune mediators have not been fully elucidated.
Healthy lungs have traditionally been believed to be sterile due to the inability to culture bacteria from lower airway samples using routine microbiological approaches. However, recent culture-independent sequencing study have identified that the lower respiratory tract contains a complex diversity of bacteria (39). Changes in this local microbial community have been associated with the exacerbation of several pulmonary disorders, such as chronic obstructive pulmonary disease, asthma, and cystic fibrosis (39). There are multiple lines of evidence that the lung microbiota is associated with lung cancer. Jin et al. published important findings of the relationship between the lung microbiota and lung cancer in a study using a genetically engineered mouse (GEM) model (40). They found that the local microbiota, such as Herbaspirillum and Sphingomonadaceae, which are associated with tumor growth, could promote inflammation and cancer progression via lung-resident γδ T cells. Depleting the microbiota or inhibiting T cell activity or their downstream effector molecules effectively suppressed lung cancer development. In addition, Tsay et al. presented that the signature of lower airway dysbiosis, an imbalance between the types of organisms present in a person’s natural microflora, was most prevalent in the group of lung cancer patients with stage IIIB–IV tumor node metastasis and was associated with poor prognosis, as shown by decreased survival among subjects with early-stage disease. They also described that a lower airway microbiota signature was associated with upregulation of the IL-17, PI3K, MAPK, and ERK pathways in the airway transcriptome, and Veillonella parvula was the most abundant taxon driving this association (41).
Human lung microbiome and mouse studies revealed that the most common bacterial genera present in the lung microbiome included Staphylococcus (~15%), Streptococcus (~15%), and Lactobacillus (~15%), along with the family Pasteurellaceae (~10%) (40,42,43). However, in the lungs of cancer patients, the total bacterial burden is significantly increased, and several bacterial taxa, including Herbaspirillum and Sphingomonadaceae, were significantly more abundant in the lungs of cancer patients than in healthy lungs (40).
Intratumoral microbiota
It is recognized that bacteria can be detected within solid tumors. To prove this concept, Nejman et al. evaluated 1,526 tumors of seven human cancer types (bone, brain, lung, melanoma, ovary, and pancreas) and extracted bacterial DNA, RNA, and lipopolysaccharide (44). They revealed that the intratumor bacteria are mostly intracellular and are present in both cancer and immune cells.
The microbiota has been similarly detected in genetically engineered de novo mouse models of lung and pancreatic cancer, suggesting that the tumor microbiome is functionally involved as a driver of tumor-promoting inflammation and malignant tumor progression (40). The live microflora appears to have a suppressive effect on local antitumor immunity to the TME. Cancer-specific mechanisms of action of microorganisms in tumors have been reported. Beyond lung cancer, there have been reports of secreted genotoxin-mediated mutagenesis in the gastrointestinal tract and urinary tract, chemotherapy resistance due to microbial metabolism and fungal activation of the host C3 complement cascade leading to greater tumor growth in pancreatic cancer, indirect amplification of cancer cell autophagy in intestinal cancer, and tumor upregulation of the levels of matrix metalloproteinases in breast cancer. In lung cancer, inflammation mediated by CagA and IL-17-producing γδ T cells and metastasis due to decreased tumor immunosurveillance have been reported (40,45).
Microbiota and lung cancer treatment
The role of the GM in immune checkpoint blockade (ICB)
The activity of the GM has many effects on host physiology, including on the development and regulation of immune responses. Recently, multiple publications have indicated that the microbiota can specifically influence the outcome of cancer immunotherapy (46-49). They demonstrate that differential GM signatures exist in patients who respond to treatment and that these favorable signatures are associated with enhanced systemic immunity and increased levels of intratumoral immune infiltrates. The mechanisms through which the GM influences the response to immunotherapy have been investigated in preclinical and clinical studies. The data suggest that gut microbes may impact antitumor immunity via several mechanisms, including the interaction of microbial components or products [such as pathogen-associated molecular patterns (PAMPS)] with antigen-presenting cells (APCs) and innate effectors [via pattern-recognition receptors (PRRs) such as Toll-like receptors (TLRs)], which can help prime the adaptive immune response; induction of cytokine production by APCs or lymphocytes; and even through inducing local or distant effects with microbial metabolites (50).
Vétizou et al. presented a study of a mouse model in which CTLA-4 inhibitory therapy resulted in a marked change in the abundance of microbes in the intestinal flora in mice, with a relative increase in Bacteroidales and Burkholderiales abundances and a decrease in Clostridiales abundance (47). In addition, they showed that the efficacy of anti-CTLA-4 therapy was markedly reduced in germ-free mice treated with broad spectrum antibiotics. Furthermore, oral feeding with Bacteroides fragilis in combination with either Bacteroides thetaiotaomicron or Burkholderia cepacia augmented the action of anti-CTLA-4 therapy by eliciting Th1 responses in the lymph nodes and facilitating the maturation of intratumoral DCs. In addition, Griffin et al. presented the molecular mechanisms that influence the host response to immunotherapy (51). They showed that members of the bacterial genus Enterococcus improve ICI in mouse models. Active enterococci express and secrete orthologs of the NIpc/p60 peptidoglycan hydrolase SagA that generate immune-active muropeptides. The expression of SagA in nonprotective E. faecalis was sufficient to promote the immunotherapy response, and its activity required the peptidoglycan sensor NOD2. Their data suggested that microbiota species with specialized peptidoglycan remodeling activity and muropeptide-based therapeutics may enhance cancer immunotherapy.
Hakozaki et al. presented an association between GM and ICI outcomes in NSCLC patients (52). They collected baseline pre-ICI samples and the clinical data of 70 Japanese NSCLC patients who received anti-PD-1/PD-L1 antibodies as a first-line or treatment-refractory therapy. In this research, patients who were treated with antibiotics before ICI had lower alpha diversity at baseline and underrepresentation of Ruminococcaceae UCG 13 and Agathobacter than those of patients not treated with antibiotics. For antibiotic-free patients at baseline, alpha diversity correlated with OS. Ruminococcaceae UCG 13 and Agathobacter levels were also higher in patients with better objective response rate (ORR) and progression-free survival (PFS). Ruminococcaceae UCG 13 abundance was higher in patients with better overall survival (OS) for more than 12 months. GM differences were observed between patients who experienced low- versus high-grade irAEs. The negative influence of antibiotics on the composition of the GM and identification of the bacterial repertoire in patients experiencing favorable responses to ICI was shown. Oncologists might recall that antibiotics lead to dysbiosis in the gut microbiome for cancer patients who are going to be treated with ICI. Therefore, the administration of unnecessary antibiotics should be avoided in principle. In other studies, Derosa et al. presented the correlation of fecal Akkermansia muciniphila abundance and ICI response (53). They performed shotgun-metagenomics-based microbiome profiling in a large cohort of patients with advanced NSCLC (n=338) treated with first- or second-line ICIs to prospectively validate the predictive value of fecal Akkermansia. Baseline stool Akkermansia abundance was associated with increased ORRs and OS in multivariate analyses, independent of PD-L1 expression, antibiotic treatment, and performance status. Moreover, intestinal Akkermansia was accompanied by richer commensalism, including with Eubacterium hallii and Bifidobacterium adolescentis, and a more inflamed TME in a subset of patients. However, antibiotic use coincided with a relative dominance of Akkermansia above 4.8% accompanied by the genus Clostridium, both associated with resistance to ICI. Significant differences in the relative abundance of Akkermansia may represent a potential biomarker to refine patient stratification.
The role of the GM in chemotherapy
The presence of intratumoral Gammaproteobacteria was found to be associated with resistance to gemcitabine chemotherapy in pancreatic Ducati adenocarcinoma patients (54).
Conventional cytotoxic chemotherapy is also dependent on intact immune responses, thus substantiating the notion that the GM could shape responses to these forms of therapy. For example, platinum-based chemotherapies and cyclophosphamide therapy cause the translocation of commensal bacteria (especially Gram-positive organisms such as Lactobacillus johnsonii and Enterococcus hirae) into mesenteric lymph nodes and can potentially facilitate robust stimulation of Th17 responses in the spleen and the induction of memory Th1 responses. Immune responses to cyclophosphamide have also been shown to be dependent on MyD88 and TLR signaling (55,56).
The GM and therapeutic toxicities
The GM has also been implicated in modulating the toxicity associated with cancer therapy. Much research has been done on the microbiome and chemotherapy toxicity and the potential interaction of the microbiome with ICIs, but only a few studies on the microbiome and cytotoxic drug toxicity exist.
Shen et al. found that the diversity of GM promotes the development of chemotherapy-induced mechanical hyperalgesia (57). Oxaliplatin-induced mechanical hyperalgesia was reduced in germ-free mice and mice pretreated with antibiotics. Germ-free mice did not suppress mechanical hyperalgesia. These effects appear to be mediated, in part, by TLR4 expressed on hematopoietic cells, including macrophages.
Some intestinal bacteria may be protective against the toxicity of cancer immunotherapy. For example, Bacteroidetes has been reported to be more common in patients resistant to colitis caused by ipilimumab, and a higher abundance of these taxa within the gut is also generally associated with a lower incidence of toxicity (58). Bifidobacterium can abrogate pathology in a mouse model of immunotherapy-induced colitis (59). Conversely, some bacterial taxa may also be associated with favorable responses as well as toxicity. Bacterial taxa within the Ruminococcaceae family have been reported to be associated with immunotherapy-induced colitis (59).
Modulating the composition of the microbiome
Therapeutic strategies that modulate the microbiome are currently being evaluated to enhance the ICI response or to avoid primary resistance to ICIs (60). GM modulation studies, such as those investigating fecal transplantation, composition, diet, and probiotics, showed that favorable microbiota modulation is related to increased levels of tumor infiltrating lymphocytes and specifically effector CD8+ T cells. This CD8+ T-cell infiltration is known to be associated with the enhanced intratumoral activity of Th1+ cells and DCs and a lower density of immune suppressive cells. Several studies as described below have begun investigating the manipulation of the intestinal microbiota to elicit the effects of immunotherapy. Several factors are known to manipulate the intestinal microbiota, including diet, bacterial administration, and fecal transplantation.
Diet
Dietary intake can also promote differences in microbiome composition, and deep and intensive changes in dietary regimens can significantly alter the GM in a relatively short time (61). In another study, Desai et al. found that when dietary fiber is chronically or intermittently deficient, the GM turns to mucus glycoproteins secreted by the host as a source of nutrition, and the colonic mucus barrier is eroded (62). Some groups have already begun to explore the impact of diet on the GM in cancer patients. The “BE GONE” trial (NCT02843425) is a study conducted by the M.D. Anderson Cancer Center to measure changes in the bacterial population in colorectal cancer patients after adding 1/2 cup of beans per day to their regular diet and supplementing with fiber.
The favorable safety profile, cost, and accessibility of dietary interventions could provide a simple and safe opportunity for assessing the implications of the microbiota and downstream immune manipulation in cancer patient populations.
Bacterial administration
Prebiotics will be an important factor in contemplating strategies to modulate gut microbes (63). Prebiotics comprise dietary compounds, including fibers and inulin, and they may support certain GM or modulate their functionality. The administration of bacterial consortia and designer probiotics might be a more feasible way to manipulate microorganisms in the clinical setting than manipulating the diet.
Recent study in preclinical models suggest that administration of inulin is associated with enhanced anti-humoral immune responses in melanoma (64). Several trials using probiotics in cancer patients have been initiated, with some completed. Clinical trials evaluating the potential effect of dietary modifications and prebiotics in metastatic cancer patients treated with immunotherapy are underway (NCT04552418, NCT04316520). In addition to lung cancer, several trials have been conducted in colorectal cancer, and many of these trials observe clinical course, changes in inflammatory response markers, and adverse events. For example, NCT03782428 is a completed study in which researchers investigated the effects of oral probiotics that contained six viable microorganisms of Lactobacillus and Bifidobacteria on the clinical course and inflammatory response markers in patients with colorectal cancer (65). They revealed that probiotics reduced levels of proinflammatory cytokines in colorectal cancer patients. In other cancer types, trials are underway to test whether probiotics affect the efficacy of ICIs and cytotoxic chemotherapy.
In lung cancer, several trials are manipulating the gut microbiome (Table 2). Seven clinical trials of oral probiotics are now recruiting patients (NCT03637803, NCT04601402, NCT04699721, NCT04857697, NCT04909034, NCT04924374, and NCT05094167).
Table 2
| Trial number | Patient population | Intervention | Outcomes | Status (country/region) |
|---|---|---|---|---|
| NCT02771470 | Lung cancer | Drug: probiotics | Composition of microorganisms in stool after probiotic intervention; frequency and severity of adverse effects during chemotherapy; the change of immunity and nutrition index | Completed (China) |
| NCT04056026 | Mesothelioma | Fecal microbiota transplant | Primary: PFS | Completed (United States) |
| NCT03637803 | Solid tumor, NSCLC, RCC, melanoma, bladder cancer | MRx0518 with pembrolizumab | Primary: safety and tolerability of MRx0518 in combination with pembrolizumab; clinical benefit of MRx0518 in combination with pembrolizumab; secondary: antitumor effect | Recruiting (United States) |
| NCT04601402 | Solid tumor, NSCLC, head and neck, urothelial carcinoma | GEN-001 with avelumab | Primary: safety; secondary: efficacy | Recruiting (United States) |
| NCT04699721 | NSCLC Stage III | Nivolumab + paclitaxel + carboplatin + BiFico (Bifidobacterium trifidum live powder) | Primary: safety, surgical complications; secondary: efficacy (ORR, recurrence rate, DFS, OS) | Recruiting (China) |
| NCT04857697 | Breast cancer, lung cancer | Oral probiotics | Primary: length of probiotics, adherence of probiotics, percentage of CD8+, CD4+, and T-reg cells, cytokine counts | Recruiting (China) |
| NCT04909034 | Lung cancer | MS20 | Primary: the incidence of treatment-emergent adverse event; secondary: ORR, PFS, DCR, DOR | Recruiting (Taiwan) |
| NCT04924374 | Lung cancer | Dietary supplement: microbiota transplant plus anti-PD-1 therapy | Primary: safety; secondary: efficacy (iRECIST) | Recruiting (Spain) |
| NCT05094167 | NSCLC | Lactobacillus Bifidobacterium V9 (Kex02) with combined carilizumab with platinum chemotherapy | Primary: ORR; secondary: PFS, OS | Recruiting (China) |
NSCLC, non-small cell lung cancer; RCC, renal cell carcinoma; PD-1, programmed death 1; PFS, progression-free survival; ORR, overall response rate; DFS, disease-free survival; OS, overall survival; DCR, disease control rate; DOR, duration of response; iRECIST, immune response evaluation criteria in solid tumors.
Another possible probiotic treatment is Akkp2611. In a study of the correlation between Akkermansia and ICI response (53), Derosa et al. suggested that therapeutic supplementation with lyophilized encapsulated Akkp2611 would benefit subgroups of patients not exposed to antibiotics and devoid of endogenous Akkermansia, and complex polymicrobial consortia or fecal microbial transplantation may be best suited for patients with prior antibiotic exposure.
Fecal microbiome transplantation and consortia
Fecal microbiota transplantation (FMT) is the most direct means of manipulating the microbiota, and fecal microbiota transplant formulations can be administered to recipient patients via oral administration of lyophilized tablets or packaged capsules or direct administration via colonoscopy or gastroscopy from an identified donor patient (66). This treatment is a recently developed therapy for recurrent Clostridium difficile infections and refractory inflammatory bowel diseases such as Crohn’s disease and ulcerative colitis. Clinical trials of FMT in cancer patients have just begun, but the results are highly anticipated. Autologous FMT in acute myeloid leukemia is being trialed in patients undergoing intensive care to increase the diversity of the GM and prevent dysbiosis during the treatment period. Furthermore, the application of FMT is being explored in patients receiving immunotherapy for solid tumor malignancies, particularly those treated with ICIs. A phase 1 single-center trial for metastatic melanoma patients who failed prior immunotherapy is underway wherein FMT from patients with a good response to immunotherapy is administered to refractory patients. The design of an additional trial is currently underway, and the aim is to test the hypothesis that modulation of the GM will improve the response to treatment with ICB. In the area of lung cancer research, there are several studies of the microbiota of lung cancer patients (52,53), and two trials are in preparation that will combine fecal transplantation or intestinal bacteria reconstitution therapy with ICIs (NCT04105270, NCT05008861). NCT04105270 is the randomized, active-controlled, parallel-group, double-blind, phase II trial of intravenous durvalumab (MEDI4736) and chemotherapy in combination with oral recovery microbiota therapy (RMT) or placebo in patients with untreated advanced or metastatic adenocarcinoma NSCLC; NCT05008861 is a study to evaluate the safety of FMT in the treatment of advanced NSCLC and to analyze the effect of FMT on patients’ GM and immune phenotype conducted by treating patients with locally advanced or metastatic NSCLC after primary treatment with PD-1/PDL-1 monoclonal antibodies in combination with PD-1/PDL-1 monoclonal antibody and GM reconstruction therapy (FMT and other therapies).
Consortia, a lab-produced or designed consortium of microbes, monoclonal microbial candidates, and bacterial peptides, might theoretically incorporate a probiotic-like agent with a high safety profile and may induce an FMT-like functional improvement, as these small microbial communities can work together. Clinical trials are underway to evaluate whether consortia enhance the function of the GM alone and in combination with immunotherapy (NCT03817125, NCT04208958). The consortia are either derived from donors having favorable GM signatures (NCT03817125) or engineered from preclinical models (67). In addition, investigations are underway to reconstruct gut microbial consortia via culturomics (68,69). Culturomics is an application of high-throughput culture conditions from human microbiota and uses matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) or 16S ribosomal RNA (rRNA) amplification and sequencing for the identification of growing colonies (70). However, we need to understand the potential complexities of the current scalability and the consistency in manufacturing such products.
Discussion: future directions
A new approach for cancer research in “Hallmarks of Cancer”
Every decade, Hallmarks of Cancer by Hanahan and Weinberg has been guided by cancer research to clarify the mechanism of cancer and lead to treatments (and improve cancer treatment) (71). In 2022, Hanahan published new dimensions of hallmarks of cancer (4). In this statement, eight hallmark capabilities and embodied characteristics that have been sufficiently validated over a decade are discussed. The current review incorporated four additional proposed emerging hallmarks and enabling characteristics involving “unlocking phenotypic plasticity”, “nonmutational epigenetic reprogramming”, “polymorphic microbiomes”, and “senescent cells”. In addition, microbiomes are introduced as ecosystems involving existing bacteria or fungi, and they have a profound impact on human health and diseases, including cancer phenotypes (5,72). However, Hanahan also considers that the onco-biome is the new frontier associated with multiple tissues and organs and that the microbiome is too complex regarding population dynamics and the diversity of macrobacterium to be able to clarify the role of this entity soon. Polymorphic microbiomes are considered to intersect with tumor-promoting inflammation and genomic instability and mutations. This polymorphic influence and interaction will be a quasi-independent variable in cancer development, progression, and response to cancer treatment. the reproducibility has not been guaranteed in each study over the cancer subtypes and the analytic method has not been established. In the 16S rRNA method, the single arm with a moderate sample size has been established, and in the next step, randomized control study of JCOG 2007 (jRCTs031210013) comparing platinum doublets with pembrolizumab and platinum doublets with nivolumab/ipilimumab is underway. In this sub-study (not yet registered), fecal samples will be analyzed to predict the efficacy and safety of each chemotherapy.
In addition, multiple tissue microbiomes are involved in modulating tumor phenotypes and the tumor response to immunotherapy positively or negatively. In particular, the use of antibiotics will cause dysbiosis in the gut flora and therefore have a negative effect on ICIs (52). Since maintaining the diversity of the GM is important, it has long been recognized that GM is fundamental for the function of the colon to degrade and import nutrients into the body to maintain metabolic homeostasis, and sometimes distortions in microbiota and dysbiosis can result in physiological maladies (73). The mechanisms by which microbiota impart these modulations are still being elucidated; however, two general effects are increasingly well understood for tumor promotion generally and the promotion of specific tumors. Through the interaction of the colonic epithelium with bacteria that promote mutagenesis, consequently, bacteria producing toxins and other harmful molecules influence either DNA damage or disruptions of the systems needed for maintaining genomic stability, or they stress the cells indirectly. In addition, metabolites such as butyrate from the microbiome, induce complex physiologic effects in nascent epithelial and fibroblastic cells (29,74). This process will lead to the expression of a diverse repertoire of cytokines or chemokines that can sculpt the abundance and characteristics of immune cell populations in the colon, comprising colonic epithelia and its underlying stroma and draining lymph nodes.
Additionally, some bacteria can form a protective biofilm and breach the mucus lining the colon, leading to destruction of the epithelial tight junctions between cells, which maintain the physical barrier that normally compartmentalizes the intestinal microbiome. Once bacteria invade the stroma of the gut, they induce innate and adaptive immune responses and elicit the secretion of a repertoire of cytokines and chemokines. Distinctive microbiomes in individual patients can be associated not only with prognosis but also with efficacy or resistance to ICIs by eliciting innate tumor-promoting inflammation and promoting tumor escape of adaptive immune destruction (3,5,75). In patients with melanoma who progressed during prior ICB, fecal transplantation from the responder restored the efficacy of ICB (60,76). Based on these results, the molecular mechanisms by which polymorphic microbiomes indirectly and systemically modulate tumor immunobiology have been demonstrated (77,78). Homeostasis, aging, and cancer, which have both overlapping and distinct species and abundances, are clearly associated with differences in the composition and diversity of the microbiome. In this context, research on the gut microbiome is sometimes considered to be data-driven research using data mining; nevertheless, the hypothesis of these studies is based on immunological insights that clarify the mechanism and enrich the related molecules to enhance ICB or overcome resistance to ICB with multiomics analysis suggested in the hallmarks of cancer. In this review, we have described a number of new clinical trials of diet, bacterial administration and FMT that intervene in the gut microbiome. It is hoped that some of these trials may lead to new therapies that improve the efficacy of ICIs or influence the attenuation of resistance to these drugs.
Microbiome analytics
Conventionally, the bacterial culture has been unrealistic since some bacterial species are often difficult to culture and identify. However, recent progress in microbiological approaches improved through the extraction of DNA from the GM, and the identification of specific bacterial species has prevented the need for bacterial culture and interactions with the environment and has elucidated changes in genetic pools. This metagenomic approach is based on next-generation sequencing and bioinformatics and has been a breakthrough in improving the comprehensive analysis of the whole genome of the microbiome.
Metagenomic analysis innovatively changes our understanding of microbial communities using a next-generation sequencer by enrichment with bioinformatics analysis. This approach is roughly divided into two methods: meta 16S analysis and full metagenome analysis. Meta 16S analysis specifically amplifies and sequences the 16S rRNA gene (18S in the case of eukaryotes) unique to prokaryotes from a group of genes extracted from fecal samples (79). The 16S rRNA is one of the components of the 1542-base-long prokaryotic rRNA. The 16S rRNA gene has a region where systematic mutations are highly preserved across bacterial species, which can be identified by comparison with existing databases. The limitation of 16S rRNA-based sequencing is that it will be biased depending on the specified region analyzed, and there is a potential bias for specific bacterial species as the sequence fragment that can be analyzed by the sequencer is from 100 to 500 base pairs. In addition, DNA extraction of feces is required for analysis, but reproducibility does not occur across each method, including cell crushing and vibration crushing. On the other hand, whole metagenome analysis sequences all bacterial fragmented gene sequences without selecting specific genetic regions. The disadvantage of whole metagenome analysis compared to 16S rRNA is the enormous amount of genomic data, which leads to a high burden for computational analysis with large amounts of time required as well as higher reagent costs. Therefore, current analysis of patient data is realistically available by 16S rRNA analysis targeting a specific region efficiently.
A recent study clarified the enriched species of the GM in the clinical setting, including the presence of Akkermansia, and demonstrated increased ORR and survival, although microbiota-relevant confounding factors should be considered (53). However, this first step of progress is important to show the role of GM modulation in immunotherapy. The translational approach, which is a bench to bedside approach and vice versa, should be further enhanced. In addition, data on clinical implementation of these approaches on the basis of real-world data, such as the use of antibiotics, which may have a negative impact on the clinical outcome, will be available by accumulating evidence.
Additionally, we have a question about whether cross-talk between the human immune system and the gut microbiome realistically contributes to modulating or interferes beyond immunomodulation and genomic alteration, thereby influencing tumorigenesis and progression. At present, there is evidence that specific bacterial species will directly stimulate proliferating singling via colonic epithelium and modulate immune cell growth suppression by altering tumor suppressor activity in a different compartment of the intestine (72,80). Recent investigations have accumulated evidence for the polymorphic variations in gut microbiomes or other organs that complement modulating tumors, such as tumor growth, inflammation in the TME, immune evasion, genomic instability, and treatment resistance (4).
Currently, the onco-biome is at the clinical trial phase for clinical implementation and has moved from the preclinical phase. In particular, preclinical landmark studies have demonstrated an association between specific GM and the effectiveness of immunotherapy (46-48,50,53). However, the exact mechanism by which the GM supports immunotherapy remains unclear, and how it works on immune cells or specific molecules produced by the microbiota remains unclear. A GM enriched for specific microbes or broad alpha diversity is associated with a favorable prognosis in patients with NSCLC treated with immunotherapy. To improve the efficacy of immunotherapy, it is important to conduct interventional studies with randomized controlled trials that modulate the GM.
Conclusions
The gut microbiome is currently becoming the hallmark of cancer research and has an established and critical role in regulating antitumor immunity and the response to ICB in patients with lung cancers. The microbiome is regarded as an organ that modulates cancer immunity, and it has been clarified that the specific bacterial species that are enriched can both augment and impede. It is important to study the oncobiome to enhance the efficacy of pharmacotherapy and adjuvant treatment.
Acknowledgments
The authors thank Editage (https://www.editage.jp/) for the English language review.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-299/rc
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-299/prf
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-299/coif). YO serves as an unpaid editorial board member of Translational Lung Cancer Research from January 2022 to December 2023. YO reports grants from AbbVie; personal fees from AstraZeneca, Nippon Boehringer Ingelheim, Eli Lilly, Ono Pharmaceutical, Taiho Pharmaceutical, and Takeda; grants and personal fees from Chugai. ATT has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Schmidt TSB, Raes J, Bork P. The Human Gut Microbiome: From Association to Modulation. Cell 2018;172:1198-215. [Crossref] [PubMed]
- Heintz-Buschart A, Wilmes P. Human Gut Microbiome: Function Matters. Trends Microbiol 2018;26:563-74. [Crossref] [PubMed]
- Fessler J, Matson V, Gajewski TF. Exploring the emerging role of the microbiome in cancer immunotherapy. J Immunother Cancer 2019;7:108. [Crossref] [PubMed]
- Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov 2022;12:31-46. [Crossref] [PubMed]
- Helmink BA, Khan MAW, Hermann A, et al. The microbiome, cancer, and cancer therapy. Nat Med 2019;25:377-88. [Crossref] [PubMed]
- Kichenadasse G, Miners JO, Mangoni AA, et al. Association Between Body Mass Index and Overall Survival With Immune Checkpoint Inhibitor Therapy for Advanced Non-Small Cell Lung Cancer. JAMA Oncol 2020;6:512-8. [Crossref] [PubMed]
- Takada K, Shimokawa M, Akamine T, et al. Association of Low Body Mass Index With Poor Clinical Outcomes After Resection of Non-small Cell Lung Cancer. Anticancer Res 2019;39:1987-96. [Crossref] [PubMed]
- Richtig G, Hoeller C, Wolf M, et al. Body mass index may predict the response to ipilimumab in metastatic melanoma: An observational multi-centre study. PLoS One 2018;13:e0204729. [Crossref] [PubMed]
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature 2018;553:446-54. [Crossref] [PubMed]
- Peters S, Camidge DR, Shaw AT, et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2017;377:829-38. [Crossref] [PubMed]
- Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med 2014;371:1963-71. [Crossref] [PubMed]
- Planchard D, Smit EF, Groen HJM, et al. Dabrafenib plus trametinib in patients with previously untreated BRAFV600E-mutant metastatic non-small-cell lung cancer: an open-label, phase 2 trial. Lancet Oncol 2017;18:1307-16. [Crossref] [PubMed]
- Drilon A, Oxnard GR, Tan DSW, et al. Efficacy of Selpercatinib in RET Fusion-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2020;383:813-24. [Crossref] [PubMed]
- Ramalingam SS, Vansteenkiste J, Planchard D, et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. N Engl J Med 2020;382:41-50. [Crossref] [PubMed]
- Skoulidis F, Li BT, Dy GK, et al. Sotorasib for Lung Cancers with KRAS p.G12C Mutation. N Engl J Med 2021;384:2371-81. [Crossref] [PubMed]
- Paik PK, Felip E, Veillon R, et al. Tepotinib in Non-Small-Cell Lung Cancer with MET Exon 14 Skipping Mutations. N Engl J Med 2020;383:931-43. [Crossref] [PubMed]
- Mazieres J, Drilon A, Lusque A, et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol 2019;30:1321-8. [Crossref] [PubMed]
- Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018-28. [Crossref] [PubMed]
- Postow MA, Sidlow R, Hellmann MD. Immune-Related Adverse Events Associated with Immune Checkpoint Blockade. N Engl J Med 2018;378:158-68. [Crossref] [PubMed]
- Postow MA, Hellmann MD. Adverse Events Associated with Immune Checkpoint Blockade. N Engl J Med 2018;378:1165. [Crossref] [PubMed]
- García-Castillo V, Sanhueza E, McNerney E, et al. Microbiota dysbiosis: a new piece in the understanding of the carcinogenesis puzzle. J Med Microbiol 2016;65:1347-62. [Crossref] [PubMed]
- Dai Z, Zhang J, Wu Q, et al. The role of microbiota in the development of colorectal cancer. Int J Cancer 2019;145:2032-41. [Crossref] [PubMed]
- Mima K, Nakagawa S, Sawayama H, et al. The microbiome and hepatobiliary-pancreatic cancers. Cancer Lett 2017;402:9-15. [Crossref] [PubMed]
- De Silva S, Tennekoon KH, Karunanayake EH. Interaction of Gut Microbiome and Host microRNAs with the Occurrence of Colorectal and Breast Cancer and Their Impact on Patient Immunity. Onco Targets Ther 2021;14:5115-29. [Crossref] [PubMed]
- Kwa M, Plottel CS, Blaser MJ, et al. The Intestinal Microbiome and Estrogen Receptor-Positive Female Breast Cancer. J Natl Cancer Inst 2016. J Natl Cancer Inst 2016;108:djw029. [PubMed]
- Noushi F, Spillane AJ, Uren RF, et al. High discordance rates between sub-areolar and peri-tumoural breast lymphoscintigraphy. Eur J Surg Oncol 2013;39:1053-60. [Crossref] [PubMed]
- Boursi B, Mamtani R, Haynes K, et al. Recurrent antibiotic exposure may promote cancer formation--Another step in understanding the role of the human microbiota? Eur J Cancer 2015;51:2655-64. [Crossref] [PubMed]
- Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 2020;580:269-73. [Crossref] [PubMed]
- Okumura S, Konishi Y, Narukawa M, et al. Gut bacteria identified in colorectal cancer patients promote tumourigenesis via butyrate secretion. Nat Commun 2021;12:5674. [Crossref] [PubMed]
- Qiu Q, Lin Y, Ma Y, et al. Exploring the Emerging Role of the Gut Microbiota and Tumor Microenvironment in Cancer Immunotherapy. Front Immunol 2020;11:612202. [Crossref] [PubMed]
- Peled JU, Gomes ALC, Devlin SM, et al. Microbiota as Predictor of Mortality in Allogeneic Hematopoietic-Cell Transplantation. N Engl J Med 2020;382:822-34. [Crossref] [PubMed]
- Schluter J, Peled JU, Taylor BP, et al. The gut microbiota is associated with immune cell dynamics in humans. Nature 2020;588:303-7. [Crossref] [PubMed]
- Staffas A, Burgos da Silva M, Slingerland AE, et al. Nutritional Support from the Intestinal Microbiota Improves Hematopoietic Reconstitution after Bone Marrow Transplantation in Mice. Cell Host Microbe 2018;23:447-457.e4. [Crossref] [PubMed]
- Kim OY, Park HT, Dinh NTH, et al. Bacterial outer membrane vesicles suppress tumor by interferon-gamma-mediated antitumor response. Nat Commun 2017;8:626. [Crossref] [PubMed]
- Mager LF, Burkhard R, Pett N, et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 2020;369:1481-9. [Crossref] [PubMed]
- Zeng Q, Michael IP, Zhang P, et al. Synaptic proximity enables NMDAR signalling to promote brain metastasis. Nature 2019;573:526-31. [Crossref] [PubMed]
- Venkatesh HS, Morishita W, Geraghty AC, et al. Electrical and synaptic integration of glioma into neural circuits. Nature 2019;573:539-45. [Crossref] [PubMed]
- Muller PA, Matheis F, Schneeberger M, et al. Microbiota-modulated CART+ enteric neurons autonomously regulate blood glucose. Science 2020;370:314-21. [Crossref] [PubMed]
- Dickson RP, Erb-Downward JR, Martinez FJ, et al. The Microbiome and the Respiratory Tract. Annu Rev Physiol 2016;78:481-504. [Crossref] [PubMed]
- Jin C, Lagoudas GK, Zhao C, et al. Commensal Microbiota Promote Lung Cancer Development via gammadelta T Cells. Cell 2019;176:998-1013 e16. [Crossref] [PubMed]
- Tsay JJ, Wu BG, Sulaiman I, et al. Lower Airway Dysbiosis Affects Lung Cancer Progression. Cancer Discov 2021;11:293-307. [Crossref] [PubMed]
- Kostic AD, Ojesina AI, Pedamallu CS, et al. PathSeq: software to identify or discover microbes by deep sequencing of human tissue. Nat Biotechnol 2011;29:393-6. [Crossref] [PubMed]
- Yun Y, Srinivas G, Kuenzel S, et al. Environmentally determined differences in the murine lung microbiota and their relation to alveolar architecture. PLoS One 2014;9:e113466. [Crossref] [PubMed]
- Nejman D, Livyatan I, Fuks G, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020;368:973-80. [Crossref] [PubMed]
- Le Noci V, Guglielmetti S, Arioli S, et al. Modulation of Pulmonary Microbiota by Antibiotic or Probiotic Aerosol Therapy: A Strategy to Promote Immunosurveillance against Lung Metastases. Cell Rep 2018;24:3528-38. [Crossref] [PubMed]
- Sivan A, Corrales L, Hubert N, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015;350:1084-9. [Crossref] [PubMed]
- Vétizou M, Pitt JM, Daillère R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015;350:1079-84. [Crossref] [PubMed]
- Frankel AE, Coughlin LA, Kim J, et al. Metagenomic Shotgun Sequencing and Unbiased Metabolomic Profiling Identify Specific Human Gut Microbiota and Metabolites Associated with Immune Checkpoint Therapy Efficacy in Melanoma Patients. Neoplasia 2017;19:848-55. [Crossref] [PubMed]
- Ripperger T, Hofmann W, Koch JC, et al. MDS1 and EVI1 complex locus (MECOM): a novel candidate gene for hereditary hematological malignancies. Haematologica 2018;103:e55-8. [Crossref] [PubMed]
- Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018;359:104-8. [Crossref] [PubMed]
- Griffin ME, Espinosa J, Becker JL, et al. Enterococcus peptidoglycan remodeling promotes checkpoint inhibitor cancer immunotherapy. Science 2021;373:1040-6. [Crossref] [PubMed]
- Hakozaki T, Richard C, Elkrief A, et al. The Gut Microbiome Associates with Immune Checkpoint Inhibition Outcomes in Patients with Advanced Non-Small Cell Lung Cancer. Cancer Immunol Res 2020;8:1243-50. [Crossref] [PubMed]
- Derosa L, Routy B, Thomas AM, et al. Intestinal Akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small-cell lung cancer. Nat Med 2022;28:315-24. [Crossref] [PubMed]
- Geller LT, Barzily-Rokni M, Danino T, et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017;357:1156-60. [Crossref] [PubMed]
- Iida N, Dzutsev A, Stewart CA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013;342:967-70. [Crossref] [PubMed]
- Viaud S, Saccheri F, Mignot G, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013;342:971-6. [Crossref] [PubMed]
- Shen S, Lim G, You Z, et al. Gut microbiota is critical for the induction of chemotherapy-induced pain. Nat Neurosci 2017;20:1213-6. [Crossref] [PubMed]
- Dubin K, Callahan MK, Ren B, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun 2016;7:10391. [Crossref] [PubMed]
- Chaput N, Lepage P, Coutzac C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol 2017;28:1368-79. [Crossref] [PubMed]
- Baruch EN, Youngster I, Ben-Betzalel G, et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2021;371:602-9. [Crossref] [PubMed]
- Tanes C, Bittinger K, Gao Y, et al. Role of dietary fiber in the recovery of the human gut microbiome and its metabolome. Cell Host Microbe 2021;29:394-407.e5. [Crossref] [PubMed]
- Desai MS, Seekatz AM, Koropatkin NM, et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 2016;167:1339-1353.e21. [Crossref] [PubMed]
- McQuade JL, Daniel CR, Helmink BA, et al. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol 2019;20:e77-91. [Crossref] [PubMed]
- Li Y, Elmén L, Segota I, et al. Prebiotic-Induced Anti-tumor Immunity Attenuates Tumor Growth. Cell Rep 2020;30:1753-1766.e6. [Crossref] [PubMed]
- Zaharuddin L, Mokhtar NM, Muhammad Nawawi KN, et al. A randomized double-blind placebo-controlled trial of probiotics in post-surgical colorectal cancer. BMC Gastroenterol 2019;19:131. [Crossref] [PubMed]
- Smits LP, Bouter KE, de Vos WM, et al. Therapeutic potential of fecal microbiota transplantation. Gastroenterology 2013;145:946-53. [Crossref] [PubMed]
- Tanoue T, Morita S, Plichta DR, et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 2019;565:600-5. [Crossref] [PubMed]
- Lagier JC, Khelaifia S, Alou MT, et al. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol 2016;1:16203. [Crossref] [PubMed]
- Fischbach MA. Microbiome: Focus on Causation and Mechanism. Cell 2018;174:785-90. [Crossref] [PubMed]
- Lagier JC, Hugon P, Khelaifia S, et al. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev 2015;28:237-64. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Dzutsev A, Badger JH, Perez-Chanona E, et al. Microbes and Cancer. Annu Rev Immunol 2017;35:199-228. [Crossref] [PubMed]
- Thomas S, Izard J, Walsh E, et al. The Host Microbiome Regulates and Maintains Human Health: A Primer and Perspective for Non-Microbiologists. Cancer Res 2017;77:1783-812. [Crossref] [PubMed]
- Salvi PS, Cowles RA. Butyrate and the Intestinal Epithelium: Modulation of Proliferation and Inflammation in Homeostasis and Disease. Cells 2021;10:1775. [Crossref] [PubMed]
- Zitvogel L, Ma Y, Raoult D, et al. The microbiome in cancer immunotherapy: Diagnostic tools and therapeutic strategies. Science 2018;359:1366-70. [Crossref] [PubMed]
- Davar D, Dzutsev AK, McCulloch JA, et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 2021;371:595-602. [Crossref] [PubMed]
- Ansaldo E, Belkaid Y. How microbiota improve immunotherapy. Science 2021;373:966-7. [Crossref] [PubMed]
- Ansaldo E, Farley TK, Belkaid Y. Control of Immunity by the Microbiota. Annu Rev Immunol 2021;39:449-79. [Crossref] [PubMed]
- Chiu CY, Miller SA. Clinical metagenomics. Nat Rev Genet 2019;20:341-55. [Crossref] [PubMed]
- Kadosh E, Snir-Alkalay I, Venkatachalam A, et al. The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature 2020;586:133-8. [Crossref] [PubMed]


