
Targeted therapies for KRAS-mutant non-small cell lung cancer: from preclinical studies to clinical development—a narrative review
Introduction
The treatment management of patients with non-small cell lung cancer (NSCLC) has profoundly improved over the last years, thanks to the possibility of exploiting an increasing number of innovative targeted therapies and immunotherapies. In current clinical practice, molecular characterization of advanced lung tumors at baseline is a necessary step to select the most appropriate first-line therapeutic strategy, as stated by international guidelines, consisting of targeted agents in those patients harboring oncogenic alterations, including mutations of epidermal growth factor receptor (EGFR), v-raf murine sarcoma viral oncogene homolog B1 (BRAF) or rearrangements of protein kinase B (ALK) or c-ros oncogene 1 (ROS1) (1-3). For those patients without driver alterations, clinicians may offer immunotherapy alone or in combination with chemotherapy, based on programmed death ligand-1 (PD-L1) expression (4). Recently, additional genetic alterations, such as those involving Kirsten rat sarcoma viral oncogene homolog (KRAS), mesenchymal-epithelial transition (MET), human epidermal growth factor receptor 2 (HER2), rearranged during transfection (RET), and neurotrophic tyrosine receptor kinase (NTRK), have gained significant clinical relevance for the development of specific inhibitors, some of which have already been approved in pretreated NSCLC patients (2,5).
KRAS belongs to the human rat sarcoma viral oncogene homolog (RAS) gene family and is frequently mutated in a variety of cancers, including pancreatic ductal adenocarcinoma (PDAC), colorectal cancer (CRC) and NSCLC (6-9). KRAS mutations are mainly represented by single-base missense mutations, almost all clustering in three hotspots at codon 12, 13, and 61 (10,11). The distribution of KRAS mutant alleles differs across tumors. Indeed, in lung cancer, in which KRAS-activating mutation is the most prevalent oncogenic driver (20–30% of cases), the dominant substitution is G12C (glycine to cysteine), whereas G12D being the most common mutation in pancreatic and CRC (11-13). KRAS mutations result in constitutive activation of downstream signaling pathways, including the rapidly accelerated fibrosarcoma (RAF)-mitogen-activated protein kinase (MEK)-extracellular signal-regulated kinase (ERK) and the phosphatidylinositol 3-kinase (PI3K)-AKT pathway, involved in different crucial processes such as proliferation, differentiation migration and survival, thereby contributing to oncogenic transformation (14-16).
Due to the important role of KRAS protein, targeting KRAS has been considered to be challenging for many years. However, the intrinsic characteristics of RAS and the complex interactions with upstream regulator proteins and downstream effectors led to consider KRAS as an “undruggable” target in lung cancer (9). Indeed, various strategies for indirectly targeting KRAS have been tested over these last years, including the inhibition of plasma membrane location or inhibition of downstream signaling pathways, and have failed to demonstrate significant activity due to the lack of selectivity for this target (9,16-18). The discovery of covalent and selective inhibitors targeting KRAS G12C mutation renewed the interest for KRAS as a valid therapeutic target (19). Currently, two principal KRAS inhibitors, sotorasib (AMG-510) and adagrasib (MRTX-849), have been proven to be highly effective in this molecularly defined subgroup of patients, and other similar drugs are currently being tested in early-phase clinical trials. Sotorasib has received approval by the U.S. Food and Drug Administration, in May 2021, as the first treatment for patients with locally advanced or metastatic NSCLC whose tumors harbor KRAS G12C mutations and who have received at least one prior systemic therapy (20).
Herein, we describe KRAS mutations and the biology of KRAS-mutant tumors and review main preclinical studies and current available data from clinical trials on KRAS inhibitors in NSCLC patients with KRAS G12C mutation. The potential mechanisms of resistance to KRAS inhibitors and investigational therapeutic strategies to further improve clinical outcome of KRAS-mutant NSCLC patients are also outlined. We present the following article in accordance with the Narrative Review reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-639/rc).
Methods
We performed an updated literature search on the role of KRAS in NSCLC on main medical research databases and on international cancer meetings websites (see Table 1 for the search strategy summary). For clinical trials, we collected and reviewed data of both completed and ongoing studies.
Table 1
| Items | Specification |
|---|---|
| Date of search | Up to August 2022 |
| Databases and other sources searched | PubMed, Scopus and Web of Science; abstracts from ESMO, ASCO, IASLC meetings |
| Search terms used | “KRAS”, “KRAS G12C”, “KRAS and lung cancer”, “KRAS inhibitors”, “sotorasib” and “adagrasib” |
| Timeframe | 1991–2022 |
| Inclusion and exclusion criteria | Only articles in English were considered |
| Selection process | Selection and collection of data was conducted independly by MS, GC, CCS, AS, MIP (as specified in authors contributions); data were analyzed and interpreted by MS, EG, NS, RR |
EMSO, European Society for Medical Oncology; ASCO, American Society of Clinical Oncology; IASLC, International Association for the Study of Lung Cancer; KRAS, Kirsten rat sarcoma viral oncogene.
Discussion
KRAS gene
RAS gene family include Harvey RAS (HRAS), KRAS, and neuroblastoma RAS (NRAS), that share significant sequence homology and encode for proteins activating multiple signaling pathways involved in cell proliferation, differentiation and survival (15,21,22). The KRAS gene, located on chromosome 12p12.1, encodes two splice variants, KRAS4A and KRAS4B, made up of 189 and 188 amino acids, respectively (16).
RAS proteins are composed of two main domains: a catalytic domain, namely the G domain, and a hypervariable region (HVR). The G domain is highly conserved and consists of three regions: the guanosine triphosphate-binding region, as well as the switch I and II regions, and is responsible for engaging with their main downstream effectors. The HVR comprises the CAAX motif, which has a key role in plasma localization (19,23). RAS is active when localized to the cell membrane. RAS association with the plasma membrane is promoted by a series of enzymatic post-translational modifications at the C-terminal CAAX motif: prenylation of the cysteine of the CAAX by farnesyltransferase (FTase) or geranylgeranyltransferase (GGTase); cleavage of the terminal AAX amino acidic residues by RAS-converting enzyme (RCE1); and methylation of the carboxyl group of the now C-terminal farnesylcysteine by isoprenylcysteine carboxyl methyltransferase (ICMT) (24). RAS localization and trafficking is regulated by the prenyl-binding protein phosphodiesterase-δ (PDEδ), which binds to farnesylated RAS. Currently, differences in RAS isoforms have been ascribed largely to sequence differences within their C-terminal HVRs, a site at which RAS proteins are differentially lipid-modified. For example, both KRAS4A and KRAS4B require an essential farnesyl moiety, but KRAS4B contains a polybasic stretch of eight lysines and KRAS4A presents a palmitoylated cysteine and two polybasic regions, suggesting distinct mechanism of plasma membrane localization and subcellular trafficking (25,26). KRAS4B has long been viewed as the major isoform as it is ubiquitously and highly expressed in human cancers (27). However, KRAS4A was shown to be widely expressed in cancer cell lines and expressed at equivalent levels to KRAS4B in colorectal tumors (28). Moreover, the expression of KRAS4A increases tumor cell adaptability to stress, such as hypoxia. This data highlights its role in tumorigenesis and as a potential therapeutic target (16).
KRAS signaling pathway
RAS proteins are membrane-bound regulatory protein (G protein), with guanosine triphosphate hydrolase (GTPase) activity and, in basal conditions, act as molecular switches alternating between two states: the GTP-bound active state and the guanosine diphosphate (GDP)-bound inactive state. These two cellular states result from activation by guanine exchange factors (GEFs), such as son of sevenless (SOS), that catalyze the loading of GTP, and inactivation by GTP hydrolysis enhanced by GTPase-activating proteins (GAP), such as neurofibromin 1 (NF1) (15,16,29). In resting state, KRAS binds to GDP in an inactivated state, due to its intrinsic GTPase activity that hydrolyses GTP to GDP.
The transition from the inactive to the active state is determined by the activation of growth factor receptors, including the EGFR family. When the EGF ligand binds its receptor, it induces an autophosphorylation process of the intracellular domain that recruits adapter proteins such as growth factor receptor bound protein 2 (GRB2) through their SH2 domains. GRB2 in turns binds SOS1, containing the RAS GEF domain. Following this stimulus, the GEF factor interacts with the inactive form of RAS protein, linked to GDP, favoring its transition to the active form linked to GTP. The RAS protein acquires an altered conformation in switch I and II and once activated allows engagement of a large number of different proteins and activation of multiple downstream intracellular pathways including the RAF-MEK-ERK pathway, the PI3K-AKT-mammalian target of rapamycin (mTOR) pathway and ral guanine nucleotide dissociation stimulator (RALGDS), which with a cascade mechanism regulate crucial cellular processes such as proliferation, differentiation, cell migration and survival (Figure 1). KRAS can also regulate the phosphatidylinositol signaling pathway by activating phospholipase C-ε (PLCε) (16,19,30). In addition to GEFs, other proteins participate to KRAS activation, such as the Src homology phosphatase 2 (SHP2), that can act as a scaffold protein to recruits GRB2-SOS1 to the receptor. Furthermore, SHP2 can activate KRAS through dephosphorylation of some regulatory molecules, including p120GAP (19,31,32).

Interestingly, a study by Tulpule et al. revealed a novel mechanism for receptor tyrosine kinase (RTK)-mediated activation of RAS-mitogen-activated protein kinase (MAPK) signaling in cancer cells. In particular, some fusion oncoproteins involving RTKs [i.e., echinoderm microtubule-associated protein-like 4 (EML4)-ALK] assembly into membrane-free cytoplasmic protein granules, which act as subcellular platform to coordinate local activation of RAS and downstream pathway independently of the lipid membrane (33).
KRAS mutations
KRAS is the most frequently mutated of the three RAS isoforms among different cancer types, representing the 75% of RAS-mutant cancers (11). KRAS mutations are characterized by single-base missense mutations and occur more frequently at codon 12 in exon 2. Other mutations can be found at codon 13 and 61.
The frequency and type of mutation vary by tissue type, suggesting the presence of specific local factors that may conditionate RAS-dependent oncogenesis in distinct cancers (11,34). KRAS mutations represent the most prevalent genomic driver event in NSCLC, present in 25–30% of adenocarcinoma, less frequently in squamous cell carcinoma (approximately 4%) (8,11,35-37). The prevalence may be lower in Asian than Western populations (38). Among all KRAS mutations, the G12C single-nucleotide variation, which causes glycine to cysteine substitution at codon 12, is the predominant mutation in NSCLC, with an overall prevalence of approximately 13% in lung adenocarcinoma (12). In large studies, the KRAS G12C represented the 40–41% of all KRAS mutations in NSCLC patients (12,35,39), followed by G12V and G12D. Overall, KRAS mutations are largely associated with smoking history: KRAS G12C is usually found in current or former smokers, whereas KRAS G12D is more typical in nonsmoking patients (35,39-41). It has been largely demonstrated that the genomic landscape is markedly distinct in never-smokers compared to smokers. Indeed, lung cancer due to tobacco smoking is characterized by a significantly higher number of mutations per Mb and mostly by C:G→A:T transversions compared to never-smokers. Interestingly, KRAS G12C and G12V arise from C→A mutations, reflecting the mutation signature associated with tobacco exposure (42,43). Most of the activating mutations of KRAS determine a disruption of GAP-mediated GTP hydrolysis and/or GDP-GTP exchange rates, resulting in accumulation of GTP-bound active state KRAS proteins. The constitutive activation of KRAS induces activation of downstream pathways involved in multiple cellular processes, including proliferation, differentiation and survival.
Heterogeneity of KRAS-mutant lung cancer
Biochemical properties, including intrinsic GTPase activity, kinetics of nucleotide-exchange, effector interactions and cell signaling, of KRAS mutants have been extensively studied (19,44,45). As commented above, activating mutations in codons 12, 13, and 61 of RAS generally disrupt GAP-mediated GTP hydrolysis (16,45). In the study by Hunter et al., the KRAS G12C has been associated with a higher intrinsic hydrolysis rate relative to other mutants (44). This allows the KRAS G12C protein to cycle from its predominant GTP-bound state to the GDP-bound state with a half-life of about 12 minutes. Therefore, in about 1 hour, 95% the KRAS G12C protein cycles through the GDP-bound state in which it is vulnerable to attack (46). This evidence served as the rationale of using covalent inhibitors specifically binding KRAS G12C in this inactive state (16,45). However, other studies have shown different results in terms of GTP hydrolysis rates for G12C and for other mutant proteins (45,47), suggesting that the activity of GDP-bound KRAS G12C inhibitors does not exclusively rely on this biochemical property. Moreover, KRAS mutations at codons 12 and 61 have been shown to be insensitive to NF1 GAP-mediated hydrolysis (48).
Cell lines with mutations of G12C and G12V seem to have decreased levels of phosphorylated AKT and increased ras-like protooncogene (RAL) signaling activation compared to wild type cell lines, while G12D mutant cell lines showed high affinity for the PI3K-AKT pathway (49,50). In the study by Ihle et al., patients harboring KRAS G12C or KRAS G12V variants have a worse progression-free survival (PFS) compared to patients with other variants or with a wild-type KRAS (49).
It has been well established that KRAS mutations have a role in modulating the immune system by inducing regulation of tumor microenvironment (TME), thereby affecting tumor progression and anti-tumor immune response (19,51). Of note, rates of high tumor mutational burden (TMB) (≥10 mutations/Mb) and PD-L1 expression varied across KRAS mutation subtypes. KRAS G12C was the most likely to be PD-L1 positive and PD-L1 high (39). As commented above, it should also be underlined that KRAS G12C are frequent in smokers and smoking is a major contributor to TMB in lung cancer as a consequence of higher level of mutagenesis from tobacco (43).
In conclusion, the genetic and phenotypic heterogeneity of KRAS mutations may have a role in determining different clinical behavior of tumors and therapeutic vulnerabilities, including chemotherapy and immunotherapy, thus likely explaining contrasting results regarding their prognostic as well as predictive role (52). These data further reinforce the importance of mutation-specific therapeutic strategies.
KRAS co-mutations
Another reason explaining heterogeneity of mutated KRAS lung tumors are the presence of concomitant genomic alterations. Co-mutations can have a significant impact on the pathogenesis, biology, micro-environmental interactions, and therapeutic vulnerabilities of NSCLC (53).
Indeed, while other driver oncogenes are commonly found in lung cancer as mutually exclusive, lung tumors with KRAS mutation represent a heterogeneous subgroup. In particular, three clusters were identified based on the patterns of co-occurring genomic alterations: one cluster dominated by co-occurring tumor protein p53 (TP53) alterations (referred to as KP), a second cluster (KL) with co-mutations or genomic loss in serine/threonine kinase 11 (STK11)/liver kinase B1 (LKB1), that was further enriched in somatic mutations in Kelch-like ECH-associated protein 1 (KEAP1) and serine/threonine kinase (ATM), and a third cluster enriched with bi-allelic inactivation of CDKN2A/B (KC), that was defined by lack of thyroid transcription factor 1 [NKX2-1 (TTF1)] transcription factor expression (54). Of note, distinct KRAS alleles were not differentially distributed between the three clusters—with the exception of enrichment for KRAS G12D in the KC subgroup in some cohorts. The presence of co-mutations induced different tumor phenotypes and relevant biologically and therapeutically differences between the subgroups (53).
The KL subgroup is characterized by the reprogramming of oxidative metabolism mediated by the factor induced by hypoxia-inducible factor 1-α (HIF-1α) and adaptation to oxidative stress. Indeed, an expression signature of the nuclear factor erythroid 2-like 2 (NRF2) gene was significantly enriched in the KL cluster. NRF2, negatively regulated by KEAP1, regulates the expression of genes encoding enzymes involved in glycolysis and glutathione synthesis, having a role in cellular defense against oxidative stress and xenobiotics. KP and KL tumors displayed distinct patterns of immune system engagement. KP tumors were characterized by an inflammatory response and their expression profiles showed enrichment for signatures of Janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway activation and interferon signaling. There was a robust expression of several co-stimulatory (e.g., CD28) and co-inhibitory molecules, including PD-L1 and a trend towards more dense infiltration with CD3+, CD8+ and CD45RO+ populations of lymphocytes. KP tumors also displayed higher global mutation rates than KL tumors, despite similar cumulative exposure to smoking. Contrary to tumors with somatic TP53 mutations, KL tumors appeared largely “immune-inert”, with reduced density of infiltrating cytotoxic CD8+ lymphocytes. Accordingly, these tumors may be particularly amenable to different therapeutic strategies: e.g., the reliance on PD-1/PD-L1 signaling and the increased immunogenicity with a large range of neoantigens suggest a role for immune checkpoint blockade in the KP subgroup. Tumor samples from 1,078 patients with KRAS mutations were analyzed by next-generation sequencing and a high frequency of co-occurring mutations in cancer-associated pathways was found, including mutations in TP53, STK11, KEAP1, ATM, MET, and Erb-b receptor tyrosine kinase 2 (ERBB2) amplification. By using an extended next-generation sequencing (NGS) panel, rare co-occurrence of targetable mutations in EGFR (13/1,078: 1.2%) and BRAF (14/1,078: 1.3%) was also revealed. Interestingly, there was an association of specific co-mutations with distinct KRAS mutation subtypes. Indeed, patients with G12C harbored all detected ERBB2 amplifications, whereas G12V and G13X mutations frequently co-occurred with phosphatase and tensin homolog (PTEN) mutations; patients with G12D showed a high prevalence of co-occurring PDGFRA mutations and lack of EGFR mutations, as well as G12A and G13X. Patients with Q61X mutations showed the highest prevalence of BRAF mutations (55). In the study by Judd et al. (39), STK11 was more frequently mutated in KRAS-mutant than wild-type NSCLC patients, with the highest rate in G13 mutations (118/327: 36.2%) and the lowest in G12D (97/684: 14.2%). Also, KEAP1 mutations were more frequent in KRAS-mutant tumors, especially in those with G13 mutations, while TP53 mutations were more frequent in KRAS WT NSCLC (73.6%), with the highest rate among KRAS mutants in those with G12other mutations (referring to other mutations different from G12C, G12V, G12D, or G12A), and the lowest in those with Q61 mutations. Other differences in co-mutated gene include U2AF1, most frequently mutated in KRAS G12other and NF1 in KRAS G13 than in other KRAS mutation subtypes (39). In a Chinese population, almost all patients with KRAS G12C mutations (representing 14.5% of KRAS mutations) had genomic aberrations associated with the RTK/RAS pathway (56).
The pattern of co-mutations, mainly STK11/LKB1 and KEAP1, have been largely evaluated and correlated with clinical and pathological features and response to immunotherapy in NSCLC patients (57). It is also important to underline that KRAS mutations regulate a complex network of multiple pathways that may play a role in the modulation of immune response and can be also potentially exploited for targeted inhibition (58).
It has been suggested that STK11/KEAP1 mutations could have negative impact on responses to immune checkpoint inhibitors (ICIs) due to the association with a ‘cold’ immune TME, characterized by a lower PD-L1 expression and levels of tumor-infiltrating lymphocytes (TILs) (53,54). Lower responses and shorter PFS and overall survival (OS) with the use of anti-PD-1 agents were observed in patients with KRAS-mutant NSCLC harboring co-mutations in STK11/LKB1 (59). In another study, co-mutation of KRAS and KEAP1 was an independent prognostic factor, predicting shorter survival and duration of response (DOR) to initial platinum-based chemotherapy, and shorter survival with immunotherapy; conversely, STK11 and TP53 did not influence the outcomes of KRAS-mutant NSCLC patients (60). In a recent work, STK11 and KEAP1 mutations were associated with worse PFS and OS to immunotherapy among patients with KRAS-mutant but not among KRAS wild-type adenocarcinoma. Tumors harboring concomitant KRAS/STK11 and KRAS/KEAP1 mutations displayed distinct immune profiles (61).
A growing evidence of data suggest that these alterations could be prognostic rather than predictive factors and may be associated to poor outcomes also to chemotherapy and targeted therapy. Indeed, recent data deriving for subgroup analyses of randomized trials in metastatic NSCLC patients indicate that a higher clinical benefit with first-line immunotherapy or immunotherapy and chemotherapy combinations compared to chemotherapy is observed regardless of STK11 or KEAP1 mutational status (57). All the above data suggest that the predictive role of co-occurring mutations need further to be explored in clinical trials including therapeutic strategies for KRAS-mutant NSCLC patients.
Interestingly, KRAS mutations and concomitant genomic alterations could be identified and monitored during the course of treatment through cell free circulating tumor DNA (ctDNA) analysis. We have identified KRAS mutations in plasma, in ctDNA, using the Guardant360 cell free DNA (cfDNA) assay. We constructed Oncoprints based on the ctDNA gene alterations and variant allele frequency (VAF) of KRAS-mutant adenocarcinoma patients, before starting first-line treatment (platinum-based chemotherapy in the majority of patients), 2 weeks after, and at the time of progression. Patients were clustered on KRAS only, KRAS + TP53 (KP), and KRAS + STK11 (KS). Patients harboring KRAS + STK11 mutations have significantly lower PFS than patients without STK11/TP53 mutations (Figure 2). In addition, higher VAF tends to correlate with shorter PFS (62). Interestingly, a recent retrospective study demonstrated the feasibility of using ctDNA to identify KRAS G12C mutations across solid tumors. Samples from metastatic patients were tested by Guardant360 assay and mutations were most frequent in patients with NSCLC, especially with nonsquamous histology. EGFR and TP53 were found to be enriched in the KRAS G12C wild-type lung cancer while STK11 was a more common co-occurring mutation in KRAS G12C-mutant lung cancer, as well as mitogen-activated protein kinase kinase 1 (MAP2K1) and PTEN alterations. More data is needed to translate this evidence into routine clinical practice (63). Beyond STK11, KEAP1 or TP53, other concomitant mutations in genes of interest, such as SMARCA4, could negatively impact on response to ICI therapy (64).

KRAS as a therapeutic target
The intrinsic biochemical characteristics of RAS led to consider it as an “undruggable” target in lung cancer (9,19). Indeed, under physiological conditions KRAS has a picomolar affinity for GTP and there is a high intracellular concentration of this trinucleotide (65). The GTP-bound KRAS presents with no binding pockets, thus highlighting the challenge of developing competitive inhibitors directly binding to this target. As commented previously, various strategies for indirectly targeting KRAS have been tested over these last years, including the inhibition of plasma membrane location or inhibition of downstream signaling pathways, with dismal results in terms of activity (19). Recent preclinical and clinical studies have returned encouraging results for target therapy in KRAS G12C-mutant-NSCLC by development of covalent, irreversible inhibitors (16-19), that will be discussed in the next paragraphs.
KRAS G12C inhibitors
KRAS G12C is associated with unique biochemical properties conferring vulnerability to covalent attack in the GDP-bound state, particularly to the active site cysteine at codon 12 (Cys12). First small molecule compounds were designed to irreversibly target KRAS G12C and demonstrated preclinical activity in KRAS-mutated cells. In the initial study by Ostrem et al., a new allosteric binding pocket adjacent to the effector region of mutant-KRAS G12C composed largely of switch-II, termed the switch-II pocket (S-IIP), which was only accessible in the GDP-bound state, was discovered (66). This region was covalently bound by specific small molecules that have been shown to completely block SOS-catalyzed nucleotide exchange and to lock GDP-bound KRAS G12C in its inactive state (66,67) These preliminary data on specific inhibitors, provided the evidence that mutant-KRAS could be targetable, while sparing wild-type KRAS and hence with potentially lower off-target toxicity. Modification and optimization of the first compounds, led to the development of a series of mutant-specific compounds, including ARS-853 and ARS-1620 (67,68), tested in preclinical trials and more active inhibitors, including AMG510 and MRTX849, that entered in clinical studies (16-19).
Sotorasib (AMG510)
Sotorasib is the first specific inhibitor of KRAS G12C to enter clinical trials. It is an oral, small molecule, highly selective inhibitor of KRAS G12C by covalently and irreversibly binding to the S-IIP, locking KRAS in its inactive GDP-bound state and inhibiting KRAS oncogenic signaling. The inhibitor also binds to a surface groove, created by an alternative orientation of histidine at position 95 (His95), leading to enhanced interaction with KRAS G12C and improved potency approximately 10-fold [mean half-maximum inhibitory concentration (IC50) =0.09 µM] as compared to ARS-1620 (69,70).
In preclinical studies, Sotorasib showed to be able to almost totally inhibit ERK-phosphorylation, a downstream effector of KRAS, and tumor cell growth in KRAS G12C-mutant cell lines in vitro and in vivo in xenograft models (69). The drug caused durable tumor regression as a monotherapy and could be combined with cytotoxic and targeted agents to synergistically kill tumor cells. It also showed a marked impact on immune cell infiltration, which renders the TME highly sensitive to immunotherapy. The study that brought to light the clinical activity of sotorasib is CodeBreak 100, a phase I study consisting of dose escalation and expansion cohorts and evaluating the safety, the pharmacokinetics and activity of sotorasib in pre-treated patients with locally advanced or metastatic solid tumors harboring the KRAS G12C mutation (71). Sotorasib was administered orally once daily. The planned dose levels for the escalation cohorts were 180, 360, 720, and 960 mg, with two to four patients receiving treatment in each cohort. The expansion cohort opened once the recommended phase 2 dose had been determined. The patients could have received prior treatment with platinum-based combination chemotherapy, anti-PD-1/PD-L1 immunotherapy or both. A total of 129 patients were included, with a median of 3 previous lines of anticancer therapies. No dose-limiting toxic effects or treatment-related deaths were observed. Treatment-related adverse events (TRAEs) were observed in 56.6% of patients and 11.6% had grade 3 or 4 events. Diarrhea, nausea, vomiting, fatigue, and elevations of aminotransferase levels were the most common adverse events, but few patients (7%) discontinued treatment because of AEs. The dose of 960 mg administered daily was identified as the dose for the expansion cohort. Sotorasib showed encouraging anticancer activity in patients with heavily pretreated advanced solid tumors. Particularly, in the subgroup with NSCLC, an objective response was observed in 32.2% (19/59) of the patients across all dose levels and 35.3% at the target dose of 960 mg, and 88.1% had disease control. Responses were durable and the median PFS was 6.3 months (71). In the phase II portion of the trial the activity of sotorasib, administered orally at the dose of 960 mg once daily, was specifically evaluated in patients with previously treated KRAS G12C-mutated advanced NSCLC, including those with stable brain metastases (72). Among the 126 enrolled patients, 81% had received both prior platinum-based chemotherapy and a PD-1/PD-L1 inhibitor. An objective response rate was observed in 46 patients (37.1%), including 4 (3.2%) who had a complete response and 42 (33.9%) with a partial response. Disease control was observed in 80.6% of patients and the median DOR was 11.1 months. The median PFS and OS were 6.8 and 12.5 months, respectively (Table 2). TRAEs occurred in 69.8% of patients, including 19.8% and 0.8% of grade 3 and 4 events. Most common grade 3 TRAEs included diarrhea, and elevated alanine aminotransferase and aspartate aminotransferase. TRAEs led to dose modification (dose interruption, reduction, or both) in 22.2% and to the discontinuation of therapy in 7.1% of patients. Of note, an assessment of exploratory biomarkers in this study revealed that responses were observed across all subgroups defined by mutation allele frequencies, PD-L1 expression, TMB and co-occurring mutations in STK11, KEAP1 or TP53 (72). Of the patients in the PD-L1-negative group [tumor proportion score (TPS) <1%], 46% had a response, as well as 42% of the overall population of patients who could be evaluated. Regarding the co-mutation subgroups, clinical responses were observed in 50% of the patients with STK11 mutations and wild-type KEAP1, hence representing the molecular subgroup with higher response to sotorasib. Among patients harboring KEAP1 mutations, a response was seen in 23% of those in the subgroup with mutations in both STK11 and KEAP1, and 14% in the subgroup with wild-type STK11 and mutated KEAP1 (72). Based on these positive results, the FDA approved sotorasib in May 2021 as the first treatment for patients with advanced, KRAS G12C-mutant NSCLC who have received at least one prior systemic therapy (20). An analysis of the patient-reported outcomes (PROs) from this study demonstrated maintenance or improvement of global health status/quality of life (QoL), physical functioning, and the severity of key lung cancer-related symptoms (73). The 2-year follow up of the CodeBreak 100, showed durable responses with sotorasib, with a 2-year OS of 32.5%. Interestingly, sotorasib was well tolerated in the long term. A total of 70% of patients experienced any TRAE; 24% had onset of a TRAE after 1 year. Grade 3 or 4 TRAEs occurred in 21%, and one of those patients had onset (of hemolytic anemia) after 1 year. No fatal TRAEs were reported, and no TRAEs led to discontinuation of therapy after 1 year (74). These promising data led to the development of an ongoing multicenter, randomized, open-label phase III study (CodeBreak 200, NCT043037780) to evaluate the efficacy of sotorasib vs. docetaxel as second-line therapy in advanced NSCLC bearing KRAS G12C, with PFS as primary endpoint (Table 2). Another ongoing study is CodeBreak 201 (NCT04933695), a phase 2 study testing sotorasib as first-line in patients with stage IV KRAS-mutant NSCLC, whose tumors have PD-L1 TPS score <1% and/or STK11 co-mutation. The primary endpoint is overall response rate (ORR) by Response Evaluation Criteria in Solid Tumors (RECIST) 1.1. Secondary endpoints include DCR, PFS and OS. Enrollment began in January 2022 and is ongoing. The prospective phase 2 Lung-MAP S1900E (NCT04625647) substudy with sotorasib will further clarify the impact of co-mutations on the efficacy of KRAS G12C inhibitors in previously treated, non-squamous NSCLC.
Table 2
| Drug inhibitor | Clinical trial | Phase | Drug combined |
|---|---|---|---|
| GDC-6036 | NCT04449874 | Ia/Ib | Atezolizumab, cetuximab, bevacizumab, erlotinib, GDC1971, inavolisib |
| JDQ-443 | NCT04699188 | Ib/II | TNO155, tislelizumab |
| D-1553 | NCT04585035 | I/II | No |
| LY3537982 | NCT04956640 | Ia/Ib | Abemaciclib, erlotinib, pembrolizumab, temuterkib, LY3295668, cetuximab, TNO155 |
| JAB-21822 | NCT05002270 | I/II | Cetuximab |
KRAS, Kirsten rat sarcoma viral oncogene.
The phase Ib/II CodeBreak 101 (NCT041185883) master protocol is also ongoing to evaluate safety, tolerability, pharmacokinetics, and efficacy of multiple combinations of sotorasib with targeted therapies, including EGFR, MEK, SHP2, pan-ErbB, mTOR and cyclin-dependent kinase (CDK) inhibitors, as well as immunotherapy and chemotherapy, in patients with advances KRAS G12C-mutated solid tumors (Table 2).
Adagrasib (MRTX849)
Adagrasib is an oral, small-molecule, covalent inhibitor of KRAS G12C that, similarly to sotorasib, irreversibly binds to the mutant protein and locks it into its inactive GDP-bound form. Adagrasib has been optimized for favorable pharmacokinetic properties, including high oral bioavailability, long half-life (approximately 24 hours), extensive tissue distribution, and central nervous system penetration. In preclinical studies, adagrasib potently inhibited KRAS-dependent signal transduction and cancer cell viability selectively in KRAS G12C cell lines and demonstrated pronounced anti-tumor activity across multiple KRAS G12C-positive cell line- and patient-derived xenograft models (75). The drug did not affect PI3K pathway. Interestingly, no co-occurring mutations correlated with response or resistance in cell lines. However, alterations of selected proteins that regulate RTK and RAS-dependent signaling and cell cycle transition in mutant KRAS, have been associated with different sensitivity to adagrasib, suggesting their potential role as therapeutic target to complement KRAS blockade. Indeed, adagrasib exhibited synergistic effects when combined with inhibitors of the EGFR family, SHP2, mTOR, or CDK4 and CDK6. Adagrasib penetrates into CSF, which may be partially mediated by its ability to penetrate tissue as well as its inhibition of P-glycoprotein-mediated efflux, being a substrate and inhibitor of P-glycoprotein, and demonstrated tumor regression and extended survival in multiple preclinical models of brain metastasis of lung cancer (76). In the same study, the authors presented preliminary data from two patients with untreated central nervous system (CNS) metastases had cerebrospinal fluid concentrations of adagrasib above the target cellular IC50, and corresponding brain metastases regression, as assessed by imaging (76). The KRYSTAL-1, a phase I/II multiple expansion cohort trial, evaluated adagrasib in patients with pre-treated, KRAS G12C-mutant advanced solid tumors. In the I/Ib dose-finding component of this trial, no MTD was defined, and a recommended dose of 600 mg twice a day was selected for the phase II. Eight of 15 patients (53.3%) evaluable KRAS G12C-mutant NSCLC achieved a confirmed partial response, with a median DOR of 16.4 months. The median PFS was 11.1 months. Most common TRAEs (any grade) were nausea (80%), diarrhea (70%), vomiting (50%), and fatigue (45%). The most common grade 3-4 TRAE was fatigue (15%) (77). The results from a phase 2 cohort of the KRYSTAL-1, evaluating adagrasib at a dose of 600 mg orally twice daily in patients with KRAS G12C-mutated NSCLC previously treated with chemotherapy and anti-PD-1/PD-L1 therapy, have been recently published (78). A total of 116 patients were included, of whom 98.3% had previously received both chemotherapy and immunotherapy. The ORR by blinded Independent Central Review was 42.9%, with a median DOR of 8.5 months, median PFS and OS of 6.5 and 12.6 months, respectively (Table 2). Among 33 patients with previously treated brain metastases, the intracranial confirmed ORR was 33.3%. Confirmed ORR were observed across all subgroups defined by co-occurring alterations in STK11, KEAP1, TP53, and CDKN2A, and PD-L1 expression. Further analysis indicated that responses were lower for those who had STK11 wild-type with a KEAP1 co-mutation (14.3%). TRAEs were registered in 97.4% of the patients, 44.8% of grade 3 or higher, of which the most common were nausea, fatigue, increased alanine aminotransferase (ALT) and aspartate aminotransferase (AST). Eight patients (6.9%) of patients discontinued adagrasib owing to TRAEs (78). On February 2022, the FDA has accepted a new drug application (NDA) for adagrasib for the treatment of patients with NSCLC harboring the KRAS G12C mutation who have received at least one prior systemic therapy.
Other cohorts of the KRYSTAL-1 study evaluating associations between adagrasib and other drugs such as afatinib, cetuximab and inhibitors of the mTOR pathway are still ongoing. The KRYSTAL-12 (NCT04685135) is a phase III study that will evaluate the efficacy of adagrasib vs. docetaxel in previously treated patients with metastatic NSCLC and KRAS G12C mutation.
Other ongoing association studies include the phase II KRYSTAL-7 (NCT04613596), evaluating the combination of adagrasib with pembrolizumab in three different cohorts as first-line treatment (cohort 1a PDL-1 TPS <1% receiving adagrasib + pembrolizumab, cohort 1b PDL-1 TPS <1% receiving adagrasib in monotherapy and cohort 2 PD-L1 TPS ≥1% receiving adagrasib + pembrolizumab), the KRYSTAL-2 (NCT04330664), testing adagrasib with TNO155 (an inhibitor of SHP2), the KRYSTAL-14 (NCT04975256) in which adagrasib is combined with BI 1701963 (SOS1 inhibitor), and the KRYSTAL-16 (NCT05178888) of adagrasib with the CDK4/6 inhibitor palbociclib Preliminary results are eagerly expected in the coming months (Table 2).
Other strategies targeting mutant-KRAS
Several ongoing studies with new KRAS G12C allosteric inhibitors as monotherapy or in combination with other agents are currently underway. A phase Ia/b study, which started in July 2020 concerns the drug GDC-6036, being evaluated both as a single oral agent and in combination with other drugs such as cetuximab, atezolizumab, bevacizumab in advanced or metastatic solid tumors with KRAS G12C mutation, including pre-treated NSCLC. Preliminary results have been recently presented at the last World Congress on Lung Cancer (WCLC). Patients received oral GDC-6036 once daily for 21-day cycles at doses of 50 mg (n=6), 100 mg (n=5), 200 mg (n=10) and 400 mg (n=6). The drug demonstrated a confirmed ORR of 46%, with 26 confirmed PRs. Regarding the safety profile, 88.1% of patients presented TRAEs, the most common observed being nausea diarrhea, and vomiting (79).
A phase I/II study (NCT04699188) is ongoing with the drug JDQ443 as a single agent and in combination with TNO155 or tislelizumab in advanced solid tumors harboring the KRAS G12C mutation. Other drugs with KRAS G12C inhibitory activity, including D-1553, LY3537982, JAB-21822, are being evaluated in phase I/II studies (Table 3).
Table 3
| Gene | Alterations |
|---|---|
| KRAS or RAS isoforms | |
| KRAS | Mutations (C12X, G13X, Q61H, R68S, H95X, Y96X) |
| KRAS amplification | |
| NRAS | Mutations (Q61X) |
| Upstream and parallel signaling | |
| RTK | EGFR mutations; EGFR, MET, FGFR amplification; RET, FGFR, ALK fusions |
| GAP inactivation | NF1 deletion |
| Downstream signaling | |
| BRAF | Mutations (V600E, K601E, G596C) |
| MEK | Mutations (K57T/N, ∆I199-K104, ∆E102-I103) |
| PIK3CA | Mutations |
| PTEN | LOF mutations |
| MYC | Amplification |
| Other pathways | |
| IDH1/2 | Mutations (R132C, R172S) |
| Phenotypic transformation | |
| Adenocarcinoma to squamous-cell transformation | |
| EMT | |
See references (80-84) for detailed description of acquired resistance mechanisms. KRAS, Kirsten rat sarcoma viral oncogene; RAS, rat sarcoma viral oncogene homolog; NRAS, neuroblastoma RAS; RTK, receptor tyrosine kinase; EGFR, epidermal growth factor receptor; MET, mesenchymal-epithelial transition; FGFR, fibroblast growth factor receptor; RET, rearranged during transfection; ALK, protein kinase B; GAP, GTPase-activating protein; GTPase, guanosine triphosphate hydrolase; BRAF, v-raf murine sarcoma viral oncogene homolog B1; MEK, mitogen-activated protein kinase; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha; PTEN, phosphatase and tensin homolog; LOF, loss-of-function; MYC, myelocytomatosis oncogene; IDH, isocitrate dehydrogenase; EMT, epithelial-mesenchymal transition.
Since the exquisite selectivity of covalent inhibitors towards the GDP-bound state of KRAS G12C, new strategies are needed to specifically target tumors expressing other KRAS mutation subtypes as well as to overcome potential mechanism of resistance inducing an active GTP-bound state of KRAS. These include additional inhibitors targeting the active GTP-bound forms of mutant-KRAS, as well as pan-KRAS inhibitors and KRAS proteolysis targeting chimeras (PROTAC) (19,85). Using structure-based drug design, revolution medicines developed potent covalent inhibitors of KRAS G12C “ON”. These inhibitors, such as RM-018, form a tricomplex between KRAS G12C “ON” and cyclophilin A. The assembled tricomplex prevents KRAS G12C “ON” from signaling via steric blockade of RAS effector signaling. These compounds have been associated with profound antitumor activity and evidence of superior activity to KRAS G12C “OFF” inhibitors in KRAS G12C-driven preclinical models (80,86,87). Moreover, this class of drugs may be effective in overcoming some type of acquired resistance mechanisms, as demonstrated by the ability of RM-018 to bind and inhibit mutant KRAS with secondary Y96D mutation conferring resistance to different KRAS G12C inhibitors binding the GDP-state, including adagrasib, in patient-derived cancer models (80).
The MRTX1133 is designed to selectively inhibit the G12D allele in both active and inactive states and is promising due to its long half-life, potency and antitumor activity on preclinical studies (88). The V941 is a lipid nanoparticle (LNP)-formulated mRNA-based cancer vaccine targeting G12D, G12V, G13D, and G12C, with potential immunostimulatory and antineoplastic activities (89), which is being evaluated in an ongoing phase I study alone or with pembrolizumab in patients with KRAS-mutant advanced or metastatic NSCLC, CRC or pancreatic adenocarcinoma (NCT03948763).
Mechanisms of resistance to KRAS inhibitors
As for other targeted therapies, different mechanisms of intrinsic and acquired resistance to KRAS G12C inhibitors have been described, thus limiting the initial magnitude of responses and the long-term efficacy of these drugs. Mechanisms of resistance include alterations of the target (e.g., secondary mutations or amplification), activation of redundant parallel signaling pathways, and histologic or phenotypic transformation (Table 3) (17-19).
Adaptative mechanisms to targeted therapies have been described in KRAS-mutant cells. In KRAS-mutant lung and colorectal cell lines treated with MEK inhibitors, ERBB2/3 expression was associated with recovery of ERK phosphorylation downstream of KRAS, conferring resistance to treatment (90). The ERBB RTKs have been demonstrated to amplify the RAS signaling pathway and support the proliferation and progression of KRAS-mutant lung tumor cells in vitro and in vivo. The broad inhibition of the ERBB with neratinib enhanced the therapeutic benefit of MEK inhibition (91).
A rapid adaptive RAS-MAPK pathway feedback reactivation following KRAS G12C inhibition by ARS-1620 and sotorasib was observed in KRAS G12C-mutant cell lines. The feedback was driven by RTK-mediated activation of wild-type RAS, which cannot be inhibited by G12C-specific inhibitors. Of note, inhibition of SHP2, which mediates signaling from multiple RTKs to RAS, abrogated this feedback reactivation, and combined KRAS G12C/SHP2 inhibition drove sustained RAS pathway suppression and improved efficacy in vitro and in vivo (92). Adaptive resistance mechanisms to ARS-1620 involving reactivation of MAPK pathway and failure to induce PI3K-AKT pathway inactivation were identified as likely resistance events in a panel of NSCLC models bearing the KRAS G12C mutation in vitro and in vivo. A high-throughput drug combinations screening identified the G12Ci + PI3Ki combination that was effective on models resistant to single-agent ARS-1620, including patient-derived xenografts (93). Despite the unclear mechanism of PI3K, AKT, and mTOR activation in KRAS G12C-mutant cells, combined inhibition of PI3K or mTOR and KRAS has more pronounced antitumor effects than either drug alone (94).
Another recent study evaluated the effect of direct KRAS G12C inhibition at single-cell resolution. It was showed that shortly after treatment with ARS-1620, some KRAS G12C-mutant cells underwent an initial growth inhibition phase, whereas others demonstrated a rapid reactivation of KRAS-oncogenic pathway with reactivation of ERK phosphorylation and proliferation (95). This divergent response occurs because some cells produced new KRAS G12C proteins that are maintained in an active, drug-insensitive state by EGFR and aurora kinase signaling. Concomitant inhibition of KRAS G12C and EGFR signaling, either by targeting EGFR or SHP2, attenuated this adaptive reactivation of GTP-KRAS (95).
Secondary mutations affecting KRAS can impact on response to selective G12C inhibitors, for example by activating nucleotide exchange or by altering GTPase activity (68). Preclinical studies suggest differential sensitivity of different acquired mutations to specific inhibitors. In a study in vitro, Koga et al. generated a total of 142 cell clones resistant to sotorasib and adagrasib, of which 124 harbored secondary KRAS mutations potentially responsible for resistance (81). In cancer cell lines resistant to either inhibitor, 12 different secondary KRAS mutations were identified. Among these, Y96D and Y96S, affecting the SP-II, were resistant to both inhibitors and interestingly, a combination of BI-3406, a SOS1 inhibitor, and trametinib, a MEK inhibitor, showed potent activity against this resistance. Although G13D, R68M, A59S and A59T were highly resistant to sotorasib, that did not suppress ERK phosphorylation levels, they remained sensitive to adagrasib, whereas Q99L displayed the opposite behavior. Furthermore, these mechanisms were also evaluated in relation to the concentration of the drug administered. For example, in cell clones treated with sotorasib, mutations A59T, R68M and Y96D were identified after treatments at high concentrations, while mutations G13D, A59S, R68M and Q61L were identified at low concentrations. This data suggests drug-specific mutations for different binding modes of the two inhibitors and suggest a potential strategy to overcome resistance by switching from one inhibitor to the other (81).
In the clinical setting, Awad et al. evaluated potential mechanisms of acquired resistance in patients with KRAS-mutant-cancers, including NSCLC, treated with adagrasib (82). These patients underwent rebiopsy at the time of disease progression to perform histologic and genomic analyses. Next generation sequencing analysis was performed on tissue (10 patients) and/or ctDNA (32 patients). In 84% of patients the original KRAS G12C mutation was identified at time of resistance. Potential resistance mechanisms were identified in 17 of 38 patients (45%). A secondary KRAS mutation, Y96C, was identified in a patient with mutant KRAS NSCLC, resulting in a change in the drug binding pocket. Other acquired KRAS alterations found in all cancers included G12D/R/V/W, G13D, Q61H, R68S, H95D/Q/R and high-level amplification of the KRAS G12C allele. Regarding bypass mechanisms, in some patients, alterations of members of RTK-RAS-MAPK pathways were detected, including mutations in NRAS (Q61K), BRAF (V600E), MAP2K1/MEK1 (Table 3). Oncogenic fusions involving ALK, RET, BRAF, serine/threonine kinase (RAF1), and fibroblast growth factor receptor 3 (FGFR3) were also identified as well as loss-of-function mutations in NF1 and PTEN. MET amplification was identified as the only potential mechanism of acquired resistance to adagrasib in two patients, one with lung adenocarcinoma. Acquired resistance was heterogeneous since 7/17 (41%) patients, mainly with CRC, had more than one concurrent resistance mechanism. Expression of these acquired mutations in Ba/F3 cells demonstrated different sensitivity to the inhibitors. Mutations within the switch II pocket, R68S, H95D/Q/R, Y96C, conferred marked resistance to adagrasib, by blocking drug binding and preventing suppression of the RAS-MAPK pathway, while R68S and Y96C, but not H95D/Q/R, mediated resistance to sotorasib, supporting the concept that differential drug-binding mechanisms between the two inhibitors can lead to the emergence of drug-specific mutations.
Another study evaluated the spectrum of genomic alterations in ctDNA of a NSCLC after developing resistance to adagrasib. Heterogeneous resistance alterations affected KRAS, NRAS, BRAF, and MAP2K1. In particular the mutation Y96D, affecting the SP-II, was demonstrated to interfere with the drug binding and confers resistance to different KRAS G12C inhibitors, which binds the GDP-state, in patient-derived cancer models. As commented above, the drug RM-018 retained the ability to bind and inhibit this mutant KRAS (80). In the study by Zhao et al., multiple treatment-emergent alterations, including alterations in KRAS, NRAS, BRAF, EGFR, FGFR2, myelocytomatosis oncogene (MYC) and other genes, were observed across patients developing resistance to sotorasib. Of note, targeted inhibition of ERK signaling intermediates enhanced the antiproliferative effect of G12C inhibitor treatment in models with acquired RAS or BRAF mutations (83).
Similar to what happens with chronic exposure to other targeted agents, including MEK inhibitors, phenotypic transformation, including epithelial-to-mesenchymal transition (EMT), has been described as mechanisms of primary and acquired resistance to KRAS G12C inhibition. In the study by Awad et al., 2 out of 9 patients with NSCLC in whom it was not possible to identify any potential genomic alteration causing resistance, a histological transformation from adenocarcinoma to squamous carcinoma was highlighted (82). In EMT-induced cells, PI3K remained activated, despite KRAS inhibition, and was mainly regulated by IGFR-IRS1 pathway. In mouse models of acquired resistance to AMG510, the combination of the KRAS G12C inhibitor, PI3K inhibitor, and SHP2 inhibitor resulted in tumor regressions (84). FGFR signaling axis activation has been also observed in mesenchymal cells following ERK and AKT inhibition by ARS-1620 (96).
The identification of mechanisms of resistance remains of crucial importance to develop additional therapeutic strategies, including the development of KRAS inhibitors with alternative binding sites and different allele specificity, and potential effective novel combination regimens (Table 3) (17,18).
Indirect strategies of targeting KRAS-mutant tumors: from preclinical evidence to clinical studies
Combining KRAS G12C inhibitors drugs targeting upstream, downstream or parallel signaling pathways could offer the potential to maximize therapeutic efficacy and delay or overcome the development of resistance mechanisms (Figure 3) (17,18).
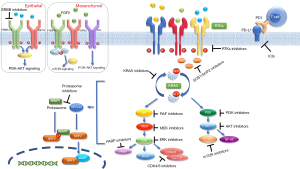
Targeting regulatory molecules
Therapeutic strategies indirectly targeting all subtypes of KRAS-mutant cancers include the inhibitors of GEF, SOS1, and SHP2, based on the evidence that RAS oncoproteins, including KRAS G12C, cycle between an inactive and active state and rely on upstream activation and nucleotide exchange to exhibit their full transforming potential (85). SHP2 inhibitors stabilize the auto-inhibited conformation of the enzyme and thereby disrupt SOS1-mediated nucleotide exchange of KRAS and may have immunomodulatory effects in T cells and macrophages to elicit antitumor immune responses (97). A phase I/II study (NCT03634982) with RMC-4630 in patients with tumors harboring RAS alterations, demonstrated in patients with KRAS G12C-mutant NSCLC a DCR of 71% (5/7) and a reduction in tumor volume in 43%, with reasonable tolerability. Clinical activity was also seen in one patient with NSCLC harboring the KRAS G12D (98). The SHP2 inhibitor TNO155 is being studied in a phase I study in advanced tumors, including KRAS G12C-mutant NSCLC (NCT03114319). Preliminary results suggest favorable pharmacokinetics and safety profile, with AEs mostly of grade 1 and 2 (99). Preclinical data suggest combined KRAS G12C and SHP2 inhibition may lead to improved clinical efficacy in KRAS G12C mutant-NSCLC (92,93).
Inhibitors of the GEF SOS1 block the interaction of SOS1 with KRAS-GDP, preventing nucleotide exchange and activation of KRAS. A synergistic effect of BAY-29 with ARS-853 was observed (100). A phase I, open-label, dose-escalation trial of the pan-KRAS SOS1 inhibitor BI 1701963 (NCT04111458) in patients with KRAS-mutated solid tumors is currently ongoing. At a preliminary analysis, the drug was generally well tolerated, and stable disease up to 18 weeks was observed in 7 of 31 patients with solid tumors harboring KRAS mutations (101).
SHP2 and SOS1 inhibitors in combination with allele-specific KRASG12C inhibitors are being tested in several ongoing clinical trials in patients with KRAS G12C mutation-positive NSCLC (NCT04185883, NCT04330664, NCT05054725, NCT04699188, NCT04973163). Recently, preliminary data from the multicenter, open-label phase Ib trial of sotorasib and RMC-4630 (NCT04185883) have been presented at the last International Association for the Study of Lung Cancer (IASLC) congress. The combination led to an investigator-assessed ORR of 27% and 50% and a DCR of 64% and 100% in pre-treated- and KRAS G12C inhibitor-naïve NSCLC patients, respectively. Treatment was safe and tolerable, with edema and diarrhea being the most common AEs. No grade 4 or fatal TRAEs and few TRAE-related discontinuations were observed.
Targeting downstream signaling molecules
RAS-RAF-MAPK pathway
Attempts to target KRAS at first centered on inhibition of the main downstream signaling pathways necessary for cell growth and proliferation, including the RAF-MEK-ERK pathway and the PI3K-AKT-mTOR pathway. Clinically co-targeting MEK and AKT signaling could be an important therapeutic strategy. Tolcher et al., (102) recommended doses of MK-2206 (AKT inhibitor) at 125 mg weekly and selumetinib (MEK1/2 inhibitor) at 100 mg once daily. Clinical responses can be seen but the durability of response to such inhibitors is curbed by incomplete cell death and development of resistance. In the basal state, a negative feedback loop has been shown from ERK to dual-specificity phosphatase (DUSP) and sprout (Spry) members, demonstrating RAS-dependent super-enhancers (103). Induction of DUSP and Sprys results in reduced phosphorylation of RTKs such as insulin-like growth factor receptor (IGF1R), and intracellular kinases, for instance, RAF and MEK. Negative regulation is lost in the presence of a MEK inhibitor. Trametinib (MEK inhibitor) induces IGF1R phosphorylation in RAS-driven rhabdomyosarcoma through loss of this negative regulation (103). In addition, AKT phosphorylation increased with trametinib. Trametinib and BMS-754807 (IGF1R inhibitor) therapy prevented ERK, AKT and IGF1R phosphorylation caused by trametinib standalone therapy (103).
Oral selumetinib with or without intravenous docetaxel was administered in previously treated patients with advanced KRAS-mutant NSCLC. Median PFS was 5.3 months in the selumetinib/docetaxel group and 2.1 months in the docetaxel group [hazard ratio (HR): 0.58, P=0.014]. The most common grade 3–4 AEs were neutropenia, 67% in the selumetinib/docetaxel group vs. 55% in the docetaxel group, and 18% febrile neutropenia registered in the selumetinib/docetaxel group (104). Other clinical studies also found limited efficacy for single-agent MAPK inhibitors, including RAF inhibitors, in KRAS-mutant tumors, suggesting combination of inhibitors targeting different molecule of the pathway, could be associated with more clinical benefit (105). The CodeBreak 101 study includes the combination of sotorasib and trametinib. The randomized, phase II, open label, RAMP-202 study (NCT04620330), is evaluating the efficacy and safety of VS-6766 (a dual RAF/MEK inhibitor) as single-agent or in combination with defactinib [a focal adhesion kinase, (FAK) inhibitor] in advanced KRAS-mutant NSCLC patients after failure of prior platinum-based chemotherapy and ICIs (106). Phase I/II studies of VS-6766 in combination with sotorasib or adagrasib in patients with KRAS G12C mutant NSCLC are ongoing (NCT05074810, NCT05375994).
SHOC2 as key mediator of response to MEK inhibitors and potential therapeutic target
Numerous research groups have used RNA-interference screening to determine various synthetic lethal targets for polytherapy therapy, such as BCL-XL, PTPN11 (encoding SHP2), YAP1, ERBB3, and FGFR1 (16). In the MEK inhibition setting in RAS-driven cancer cells, CRISPR-Cas9 loss-of-function screens using single guide RNA (sgRNA) have discovered that SHOC2 is a major regulator of KRAS-mutant cancer cell proliferation and survival after MEK inhibition (16,107). SHOC2 is a leucine-rich repeat protein that positively regulates the RAS-MAPK pathway. SHOC2 binds the catalytic subunit of PP1 (PP1c) and MRAS, resulting in membrane localization and dephosphorylation of c-Raf proto-oncogene (CRAF) at S259, thus leading to CRAF activation (17). SHOC2 regulates RTK feedback signaling in response to MEK inhibition. Upon trametinib treatment, phosphorylated-RTK arrays show that the most activated RTKs were MET/HGFR, HER3, insulin receptor (IR), and IGF1R in five KRAS-mutant cell lines (NCI-H23, A549, NCI-H2030, MIA Paca-2, and PA-TU-8902), the two latter are pancreatic cancer cells. NCI-H23 upregulates mostly MET while, for example, A549 induces RYK (107). Differential sensitivity scores from the CRISPR-MEK inhibition demonstrated that SHOC2 shows great positive correlations with several members of the RTK signaling pathway, including PTPN11, GRB2, SOS1, KRAS, BRAF, and RAF1. Knockout, suppression, or degradation of SHOC2 specifically cooperated with MEK inhibition to impair proliferation in RAS-driven cancer cells. Intriguingly, in A549 and NCI-H2030 there was a strong correspondence between the degree of sensitization mediated by SHOC2 knockdown and SHP099 (SHP2 inhibitor) sensitivity during trametinib co-treatment, leading the authors to postulate analogous functional roles for SHP2 and SHOC2 in regulating RTK-feedback signaling in response to trametinib (107). The MRAS, SHOC2 and PP1c complex regulates CRAF activation by dephosphorylation at S259, resulting in 14-3-3 displacement and increased membrane localization. Such membrane localization encourages CRAF dimerization and RAS-RAF-MAPK signaling pathway activation. Loss of SHOC2 increases the inhibitory p-S259 site of RAF1 due to trametinib treatment, averting RAF dimerization and downstream signaling to reactivate ERK1/2 (107,108). The RAF and ERK pathway activation, independent of SHOC2, is mediated by internalization of H/NRAS and CRAF, requiring FAK/p21-activated kinase (PAK)-regulated phosphorylation and cRAF activation (109). The investigators propose a model where the rapid phase of SHOC2-dependent ERK activation occurs at the plasma membrane, upon EGF stimulation, and where, upon MRAS activation, the SHOC2 complex formation leads to S259 dephosphorylation on proximal A/B/C-RAF proteins recruited by H/N/K-RAS proteins. The slow, sustained phase of ERK activation may be driven by internalization of palmitoylated RAS proteins segregated from the SHOC2 complex that remains anchored at the plasma membrane by MRAS, alongside KRAS4B. The authors suggest a model of SHOC2 dependent and independent mechanisms of ERK in anchorage-dependent/two-dimensional (2D) vs. anchorage-independent/three-dimensional (3D) conditions. In addition to SHOC2 and cRAF-dependent mechanisms at adhesion sites of the extracellular matrix, integrins activate ERK signaling, independent of SHOC2, through CRAF N-region phosphorylation by FAK/SRC and PAK kinases. In the absence of adhesion to extracellular matrix (cells in suspension or 3D), integrin-mediated ERK (and PI3K/AKT) activation is lost, and SHOC2-dependent mechanisms are more relevant to ERK activity in KRAS-mutant cells. This study sheds further light on the contribution of the SHOC2 phosphatase complex to RAF regulation and ERK pathway dynamics (109). It has recently been confirmed that the expression of SHOC2 affects sensitivity to EGFR-TKIs and EGFR-TKI/MEK inhibitor combinatory treatment. Treatment with MEK inhibitors, trametinib or selumetinib, inhibited cell proliferation when combined with osimertinib in SHOC2 depleted PC9 cells (110). Interestingly, celastrol, a pharmacologically active triterpenoid extracted from the Chinese herb Tripterygium wilfordii, binds SHOC2 and inhibits its function (111).
All the above data could suggest a role for SHOC2 inhibitors (at present not available) as single agent or better to optimize combinatory treatments for example with MEK inhibitors, as well as potential biomarker of response to SHP2 inhibitors.
PI3K-AKT-mTOR pathway
RAS-mediated activation of PI3K phosphorylates phosphatidylinositol-4,5-biphosphate (PIP2) to phosphatidylinositol-(3-5)-triphosphate (PIP3), thus activating AKT phosphorylation at T308 through 3-phosphoinositide-dependent kinase-1 (PDK1). AKT is multifunctional, activating both the mTOR complex I (mTORC1) while increasing tumorigenesis and drug resistance. Upon activation of mTORC1, its downstream targets, ribosomal protein S6 kinase (S6K), unc-51-like kinase-1 (ULK1), and eukaryotic translation initiation factor 4E (eIF4EF)-binding protein 1 (4EBP1), are phosphorylated (112). The combination of G12C and PI3K inhibitors has shown to be effective in preclinical studies including models resistant to single-agent ARS-1620 (93). The PI3K inhibitor serabelisib is being investigated in a phase I/II combination study including patients with advanced solid tumors and phosphatidylinositol-4, 5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) or KRAS mutations (NCT04073680).
Molina-Arcas et al. showed that combination of IGF1R with mTOR inhibitors caused a strong inhibition of PI3K/AKT and mTOR pathways (94). In three KRAS-mutant NSCLC cell lines (H23, H358, and H1792) inhibition of IGF1R (linsitinib) suppressed AKT phosphorylation. Linsitinib abrogated the reactivation of AKT phosphorylation produced by everolimus in both phosphosites, S473 and T308. The combination of rapalogs or mTOR kinase inhibitors with IGF1R inhibitors induced a strong inhibition of PI3K/AKT and mTOR pathways. MTOR inhibition in cells with KRAS mutations induced IGF1R and insulin receptor phosphorylation in the three KRAS cell lines. The combination of ARS1620 plus everolimus and linsitinib also reduced reactivation of RAS and ERK activation observed at 48 hours. The three-drug combination enhanced tumor regression compared to combined MEK inhibitors or ARS-1620 alone in a series of KRAS-driven mouse lung cancer models (94). These data suggest RTK/PI3K/AKT/mTOR as a target pathway for clinical investigation in KRAS-mutant tumors.
Preclinical data also suggest novel potential target to be exploited to inhibit the PI3K-AKT-mTOR pathway in KRAS-mutant NSCLC. TRIB3, a pseudo-kinase belonging to the tribbles family, inhibits AKT phosphorylation. It has been noted that arsenite therapy (113) and endoplasmic-reticulum stress (114) transcriptional activation of TRIB3 can occur through transcription factors, such as activation transcription factor 4 (ATF4)-C/EBP homologous protein (CHOP). Activation of TRIB3 suppresses AKT activity in lung and pancreatic cancer cells (114). PIERCE1, p53-induced expression in retinoblastoma (RB)-null cells 1, is a tumor-associated protein. NSCLC patients with low PIERCE1 expression have better OS and PFS rates vs. patients with high expression levels (115). PIERCE1 knockdown repressed proliferation in five of seven lung cancer cell lines (H358, H1373, H3122, H226, and HCC827). No effect was seen in PC-9 and H1299 cell lines. Also, no effect was seen in one immortalized human bronchial epithelial cell line, BEAS-2B. The increased PIERCE1 expression found in lung adenocarcinomas has a significant parallelism with KRAS mutational status (115). Growth of all KRAS-mutant cells (A549, H358, H460, and H1373) was lessened by PIERCE1 knockdown, but only in half of KRAS wild-type lung cancer cell lines. The investigators further showed in A549 cells that AKT phosphorylation at S473 was decreased in PIERCE1 knockdown, although no noticeable changes were detected in pAKT at T308 and pERK. Furthermore, it was revealed that PIERCE1 activates the AKT pathway by negative regulation of TRIB3 expression. Since TRIB3 is controlled by ATF4-CHOP transcription factors, RT-PCR further noted that PIERCE1 knockdown upregulated CHOP-responsive genes, while PIERCE1 over-expression downregulated these genes. In KRAS-mutant NSCLC, PIERCE1 negatively regulates TRIB3 and activates AKT pathway (114). Further research is warranted to understand mediators of endoplasmic reticulum stress and its negative regulators, such as eukaryotic initiation factor 5B (eIF5B), and regulation of ATF4 mRNA (116). CDK are key regulators of the cell cycle, activated downstream of KRAS. Based on preclinical findings showing synergy with KRAS G12C covalent inhibitors (69), this option could be potentially effective in the clinical setting. The CodeBreak 101 is testing the association of sotorasib with the CDK4/6 inhibitor palbociclib.
Targeting the EMT state
Solanki et al. have been able to sub-classify KRAS LUAD cell lines according to EMT state and regulatory hubs by phospho-proteomic analysis (96). A heterogenous response to ARS-1620 was observed in a panel of 8 KRAS-G12C cell lines in both 2D and 3D cultures. H358, Calu1, and H1792 cells were classified as sensitive, moderate, and resistant lines, respectively. In H358 cells 6 hours after ARS-1620 treatment, several sub-networks were activated, including a signaling hub surrounding RAF1 as a direct substrate of several kinases, such as CAMKK2, PRKCA, and PAK4, among others. It is important to note that Y1289 phosphorylation of HER3 was also observed. The ARS-1620 combination with the pan-ERBB inhibitor, afatinib, reduced cell viability in H358 cells and western blot-based signaling analysis showed longer suppression of pERK and pAKT with the combination. The data support that HER2/HER3 signaling is associated with an epithelial subtype related to co-treatment activity with a KRAS G12C inhibitor plus a pan-HER inhibitor (96). Previous studies have shown that either afatinib or neratinib (multi-ERBB inhibitors) suppresses formation of KRAS G12D-driven lung tumors and inhibition of the ERBB3 network enhances the therapeutic benefit of MEK inhibition (91,117). Neither afatinib, gefitinib nor erlotinib inhibits phosphorylation of ERBB2 and ERBB3 in A549 cell line (117).
Loss of function of FAT1, encoding a tumor suppressor, activates a Calmodulin-dependent protein kinase II (CAMK2)-CD44-SRC axis that promotes YAP1 nuclear translocation and ZEB1 expression that stimulates the mesenchymal phenotype (118). FAT1 loss induced a decrease in the total levels of EGFR and phosphorylated EGFR. CAMK2 was the kinase most frequently upstream of phospho-peptides enriched in FAT1-knockout tumor cells. At least in squamous cancer cell models, FAT1 loss of function activates a CAMK2-CD44-SRC-YAP-ZEB1 axis that promotes the expression of a mesenchymal program. FAT1-knockout cells were more resistant to afatinib and trametinib as compared to FAT1 wild-type squamous cancer cells in vitro. By contrast, FAT1-knockout tumor cells were significantly more sensitive to the SRC inhibitors, dasatinib and saracatinib, and the CAMK2 inhibitor, KN93, as compared to FAT1 wild-type tumor cells. It is of future interest to assess by NGS the FAT1 mutational status in lung adenocarcinoma in order to determine whether it can be associated with KRAS mutations. The model described, depicting that FAT1-mutated cancer cells are sensitive to CAMK2 and SRC inhibition and to resistance to EGFR and MEK inhibition (118), mirrors the relevance of EMT in KRAS cancer cell lines.
In mesenchymal phenotype KRAS, H1792 cells demonstrated modest response to dual pan-HER/KRAS-G12C inhibition. Phospho-proteomic analysis showed increased phosphorylation of FGFR1 and other components of the FGFR signaling axis with increased phosphorylation of phospholipase C gamma 1 (PLCG1), ABL1, and CDK5. Also, increased phosphorylation of the FGFR adaptor protein 2 (FRS2) was seen in H1792 following ARS-1620 treatment. Such results reinforce previous observations of the key function of FGFR1 signaling in mesenchymal type tumors and the synergistic effects of FGFR inhibition with either MEK or KRAS-G12C inhibitors, respectively (96,119). The same study by Solanki et al. identified anexelekto (AXL) receptor mediating adaptive rewiring to KRAS G12C inhibition in Calu 1 cell line. FGFR1 and AXL represent two subtypes with mesenchymal models (96).
Conclusions
The discovery and demonstration of clinical efficacy of selective drugs for KRAS G12C has witnessed the unlocked potential of targeted inhibition of this oncogene and has provided new therapeutic opportunities for this molecularly-defined subgroup of NSCLC patients. However, the complexity of the biology of KRAS mutations and the presence of intrinsic and acquired resistance mechanisms, suggest the need for more basic, translational and clinical research to optimize the use of these agents and further improve patients’ outcomes. The high frequency of co-mutations, such as those involving LKB1/STK11, KEAP1, and TP53, should be taken into consideration when developing therapeutic strategies for KRAS-mutant tumors since, as commented in the text, preclinical and clinical data suggest differential sensitivity to KRAS inhibitors and to immunotherapy. Refinement of biomarker (PD-L1, commutations) selection for use of KRAS G12C inhibitors in the frontline setting and combination strategies will be of interest, particularly with immunotherapy, since the optimal sequencing of KRAS G12C inhibitors and immunotherapy remains to be determined. The established effects of KRAS signaling on TME and immune response (19) have generated a great expectancy from ongoing combination studies with ICIs, that may synergize with G12C inhibitors to enhance CD8+ T-cell infiltration and inhibit tumor growth. Recently, a first report on the safety and efficacy of sotorasib in combination with pembrolizumab or atezolizumab in advanced KRAS G12C-mutated NSCLC demonstrated durable objective responses. However, the combination led to a higher incidence of grade 3–4 TRAEs than previously observed with sotorasib in monotherapy, primarily liver enzyme elevations. Low rates of hepatotoxicity were observed with a lead-in strategy of sotorasib monotherapy followed by the combination (120). As for other combination regimens, it would be crucial to carefully assess specific toxicities of anti-KRAS G12C drugs when associated with immunotherapy or other therapies. Novel therapeutic strategies to target KRAS-mutant tumors have been developed and deserve to be further evaluated, and these include cancer vaccines, adoptive cell therapy and inhibitors of metabolic pathway and autophagy blockade (e.g., through ULK inhibitors) (16,19,85). It remains crucial to characterize the complete molecular profile of KRAS-mutant lung tumors to identify potential determinants of response or resistance to the selective inhibitors. In the context of the acquired resistance, performing studies on serial tissue biopsy, when feasible, as well as on liquid biopsy that is easier to repeat over time, may provide important insights on genomic changes of cancer cells under selective pressure of G12C inhibitors that can also be exploited as therapeutic targets.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-639/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-639/coif). RR serves as an unpaid editorial board member of Translational Lung Cancer Research from June 2022 to May 2023. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hanna NH, Robinson AG, Temin S, et al. Therapy for Stage IV Non-Small-Cell Lung Cancer With Driver Alterations: ASCO and OH (CCO) Joint Guideline Update. J Clin Oncol 2021;39:1040-91. [Crossref] [PubMed]
- Singh N, Temin S, Baker S Jr, et al. Therapy for Stage IV Non-Small-Cell Lung Cancer With Driver Alterations: ASCO Living Guideline. J Clin Oncol 2022;40:3310-22. [Crossref] [PubMed]
- Lindeman NI, Cagle PT, Aisner DL, et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment With Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol 2018;13:323-58. [Crossref] [PubMed]
- Singh N, Temin S, Baker S Jr, et al. Therapy for Stage IV Non-Small-Cell Lung Cancer Without Driver Alterations: ASCO Living Guideline. J Clin Oncol 2022;40:3323-43. [Crossref] [PubMed]
- Santarpia M, Massafra M, Gebbia V, et al. A narrative review of MET inhibitors in non-small cell lung cancer with MET exon 14 skipping mutations. Transl Lung Cancer Res 2021;10:1536-56. [Crossref] [PubMed]
- Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017;32:185-203.e13. [Crossref] [PubMed]
- Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Cox AD, Fesik SW, Kimmelman AC, et al. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 2014;13:828-51. [Crossref] [PubMed]
- Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer Res 2012;72:2457-67. [Crossref] [PubMed]
- Prior IA, Hood FE, Hartley JL. The Frequency of Ras Mutations in Cancer. Cancer Res 2020;80:2969-74. [Crossref] [PubMed]
- Biernacka A, Tsongalis PD, Peterson JD, et al. The potential utility of re-mining results of somatic mutation testing: KRAS status in lung adenocarcinoma. Cancer Genet 2016;209:195-8. [Crossref] [PubMed]
- Li S, Balmain A, Counter CM. A model for RAS mutation patterns in cancers: finding the sweet spot. Nat Rev Cancer 2018;18:767-77. [Crossref] [PubMed]
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer 2003;3:11-22. [Crossref] [PubMed]
- Simanshu DK, Nissley DV, McCormick F. RAS Proteins and Their Regulators in Human Disease. Cell 2017;170:17-33. [Crossref] [PubMed]
- Moore AR, Rosenberg SC, McCormick F, et al. RAS-targeted therapies: is the undruggable drugged? Nat Rev Drug Discov 2020;19:533-52. [Crossref] [PubMed]
- Kim D, Xue JY, Lito P. Targeting KRAS(G12C): From Inhibitory Mechanism to Modulation of Antitumor Effects in Patients. Cell 2020;183:850-9. [Crossref] [PubMed]
- Rosell R, Aguilar A, Pedraz C, et al. KRAS inhibitors, approved. Nat Cancer 2021;2:1254-6. [Crossref] [PubMed]
- Huang L, Guo Z, Wang F, et al. KRAS mutation: from undruggable to druggable in cancer. Signal Transduct Target Ther 2021;6:386. [Crossref] [PubMed]
- U.S. Food and Drug Administration. FDA Approves First Targeted Therapy for Lung Cancer Mutation Previously Considered Resistant to Drug Therapy. 2021. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-targeted-therapy-lung-cancer-mutation-previously-considered-resistant-drug
- Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer 2003;3:459-65. [Crossref] [PubMed]
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer 2011;11:761-74. [Crossref] [PubMed]
- Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature 1991;349:117-27. [Crossref] [PubMed]
- Cox AD, Der CJ, Philips MR. Targeting RAS Membrane Association: Back to the Future for Anti-RAS Drug Discovery? Clin Cancer Res 2015;21:1819-27. [Crossref] [PubMed]
- Rásó E. Splice variants of RAS-translational significance. Cancer Metastasis Rev 2020;39:1039-49. [Crossref] [PubMed]
- Busquets-Hernández C, Triola G. Palmitoylation as a Key Regulator of Ras Localization and Function. Front Mol Biosci 2021;8:659861. [Crossref] [PubMed]
- Plowman SJ, Berry RL, Bader SA, et al. K-ras 4A and 4B are co-expressed widely in human tissues, and their ratio is altered in sporadic colorectal cancer. J Exp Clin Cancer Res 2006;25:259-67. [PubMed]
- Tsai FD, Lopes MS, Zhou M, et al. K-Ras4A splice variant is widely expressed in cancer and uses a hybrid membrane-targeting motif. Proc Natl Acad Sci U S A 2015;112:779-84. [Crossref] [PubMed]
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell 2007;129:865-77. [Crossref] [PubMed]
- Drosten M, Barbacid M. Targeting the MAPK Pathway in KRAS-Driven Tumors. Cancer Cell 2020;37:543-50. [Crossref] [PubMed]
- Ruess DA, Heynen GJ, Ciecielski KJ, et al. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat Med 2018;24:954-60. [Crossref] [PubMed]
- Bunda S, Burrell K, Heir P, et al. Inhibition of SHP2-mediated dephosphorylation of Ras suppresses oncogenesis. Nat Commun 2015;6:8859. [Crossref] [PubMed]
- Tulpule A, Guan J, Neel DS, et al. Kinase-mediated RAS signaling via membraneless cytoplasmic protein granules. Cell 2021;184:2649-2664.e18. [Crossref] [PubMed]
- Hobbs GA, Der CJ, Rossman KL. RAS isoforms and mutations in cancer at a glance. J Cell Sci 2016;129:1287-92. [Crossref] [PubMed]
- Dogan S, Shen R, Ang DC, et al. Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin Cancer Res 2012;18:6169-77. [Crossref] [PubMed]
- Jordan EJ, Kim HR, Arcila ME, et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov 2017;7:596-609. [Crossref] [PubMed]
- El Osta B, Behera M, Kim S, et al. Characteristics and Outcomes of Patients With Metastatic KRAS-Mutant Lung Adenocarcinomas: The Lung Cancer Mutation Consortium Experience. J Thorac Oncol 2019;14:876-89. [Crossref] [PubMed]
- Dearden S, Stevens J, Wu YL, et al. Mutation incidence and coincidence in non small-cell lung cancer: meta-analyses by ethnicity and histology (mutMap). Ann Oncol 2013;24:2371-6. [Crossref] [PubMed]
- Judd J, Abdel Karim N, Khan H, et al. Characterization of KRAS Mutation Subtypes in Non-small Cell Lung Cancer. Mol Cancer Ther 2021;20:2577-84. [Crossref] [PubMed]
- Yu HA, Sima CS, Shen R, et al. Prognostic impact of KRAS mutation subtypes in 677 patients with metastatic lung adenocarcinomas. J Thorac Oncol 2015;10:431-7. [Crossref] [PubMed]
- Redig AJ, Chambers ES, Lydon CA, et al. Genomic complexity in KRAS mutant non-small cell lung cancer (NSCLC) from never/light-smokers v smokers. J Clin Oncol 2016;34:abstr 9087.
- Govindan R, Ding L, Griffith M, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012;150:1121-34. [Crossref] [PubMed]
- Tan C, Mandell JD, Dasari K, et al. Heavy mutagenesis by tobacco leads to lung adenocarcinoma tumors with KRAS G12 mutations other than G12D, leading KRAS G12D tumors-on average-to exhibit a lower mutation burden. Lung Cancer 2022;166:265-9. [Crossref] [PubMed]
- Hunter JC, Manandhar A, Carrasco MA, et al. Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Mol Cancer Res 2015;13:1325-35. [Crossref] [PubMed]
- Burge RA, Hobbs GA. Not all RAS mutations are equal: A detailed review of the functional diversity of RAS hot spot mutations. Adv Cancer Res 2022;153:29-61. [Crossref] [PubMed]
- McCormick F. Sticking it to KRAS: Covalent Inhibitors Enter the Clinic. Cancer Cell 2020;37:3-4. [Crossref] [PubMed]
- Moghadamchargari Z, Huddleston J, Shirzadeh M, et al. Intrinsic GTPase Activity of K-RAS Monitored by Native Mass Spectrometry. Biochemistry 2019;58:3396-405. [Crossref] [PubMed]
- Rabara D, Tran TH, Dharmaiah S, et al. KRAS G13D sensitivity to neurofibromin-mediated GTP hydrolysis. Proc Natl Acad Sci U S A 2019;116:22122-31. [Crossref] [PubMed]
- Ihle NT, Byers LA, Kim ES, et al. Effect of KRAS oncogene substitutions on protein behavior: implications for signaling and clinical outcome. J Natl Cancer Inst 2012;104:228-39. [Crossref] [PubMed]
- Muñoz-Maldonado C, Zimmer Y, Medová M. A Comparative Analysis of Individual RAS Mutations in Cancer Biology. Front Oncol 2019;9:1088. [Crossref] [PubMed]
- Cullis J, Das S, Bar-Sagi D. Kras and Tumor Immunity: Friend or Foe? Cold Spring Harb Perspect Med 2018;8:a031849. [Crossref] [PubMed]
- Ferrer I, Zugazagoitia J, Herbertz S, et al. KRAS-Mutant non-small cell lung cancer: from biology to therapy. Lung Cancer 2018;124:53-64. [Crossref] [PubMed]
- Skoulidis F, Heymach JV. Co-occurring genomic alterations in non-small-cell lung cancer biology and therapy. Nat Rev Cancer 2019;19:495-509. [Crossref] [PubMed]
- Skoulidis F, Byers LA, Diao L, et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015;5:860-77. [Crossref] [PubMed]
- Scheffler M, Ihle MA, Hein R, et al. K-ras Mutation Subtypes in NSCLC and Associated Co-occuring Mutations in Other Oncogenic Pathways. J Thorac Oncol 2019;14:606-16. [Crossref] [PubMed]
- Loong HH, Du N, Cheng C, et al. KRAS G12C mutations in Asia: a landscape analysis of 11,951 Chinese tumor samples. Transl Lung Cancer Res 2020;9:1759-69. [Crossref] [PubMed]
- Di Federico A, De Giglio A, Parisi C, et al. STK11/LKB1 and KEAP1 mutations in non-small cell lung cancer: Prognostic rather than predictive? Eur J Cancer 2021;157:108-13. [Crossref] [PubMed]
- Santarpia M, Aguilar A, Chaib I, et al. Non-Small-Cell Lung Cancer Signaling Pathways, Metabolism, and PD-1/PD-L1 Antibodies. Cancers (Basel) 2020;12:1475. [Crossref] [PubMed]
- Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov 2018;8:822-35. [Crossref] [PubMed]
- Arbour KC, Jordan E, Kim HR, et al. Effects of Co-occurring Genomic Alterations on Outcomes in Patients with KRAS-Mutant Non-Small Cell Lung Cancer. Clin Cancer Res 2018;24:334-40. [Crossref] [PubMed]
- Ricciuti B, Arbour KC, Lin JJ, et al. Diminished Efficacy of Programmed Death-(Ligand)1 Inhibition in STK11- and KEAP1-Mutant Lung Adenocarcinoma Is Affected by KRAS Mutation Status. J Thorac Oncol 2022;17:399-410. [Crossref] [PubMed]
- Bracht J, Karachaliou N, Lanman RB, et al. Tracking plasma KRAS mutations (mu) in lung adenocarcinoma (LUAC) patients (p) and branching evolution. J Clin Oncol 2019;37:abstr 9055.
- Thein KZ, Biter AB, Banks KC, et al. Identification of KRAS(G12C) Mutations in Circulating Tumor DNA in Patients With Cancer. JCO Precis Oncol 2022;6:e2100547. [Crossref] [PubMed]
- Liu L, Ahmed T, Petty WJ, et al. SMARCA4 mutations in KRAS-mutant lung adenocarcinoma: a multi-cohort analysis. Mol Oncol 2021;15:462-72. [Crossref] [PubMed]
- Goody RS, Frech M, Wittinghofer A. Affinity of guanine nucleotide binding proteins for their ligands: facts and artefacts. Trends Biochem Sci 1991;16:327-8. [Crossref] [PubMed]
- Ostrem JM, Peters U, Sos ML, et al. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 2013;503:548-51. [Crossref] [PubMed]
- Patricelli MP, Janes MR, Li LS, et al. Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer Discov 2016;6:316-29. [Crossref] [PubMed]
- Lito P, Solomon M, Li LS, et al. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science 2016;351:604-8. [Crossref] [PubMed]
- Canon J, Rex K, Saiki AY, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019;575:217-23. [Crossref] [PubMed]
- Lanman BA, Allen JR, Allen JG, et al. Discovery of a Covalent Inhibitor of KRAS(G12C) (AMG 510) for the Treatment of Solid Tumors. J Med Chem 2020;63:52-65. [Crossref] [PubMed]
- Hong DS, Fakih MG, Strickler JH, et al. KRAS(G12C) Inhibition with Sotorasib in Advanced Solid Tumors. N Engl J Med 2020;383:1207-17. [Crossref] [PubMed]
- Skoulidis F, Li BT, Dy GK, et al. Sotorasib for Lung Cancers with KRAS p.G12C Mutation. N Engl J Med 2021;384:2371-81. [Crossref] [PubMed]
- Spira AI, Wilson FH, Shapiro G, et al. Patient-reported outcomes (PRO) from the phase 2 CodeBreaK 100 trial evaluating sotorasib in KRAS p. G12C mutated non-small cell lung cancer (NSCLC). J Clin Oncol 2021;39:abstr 9057.
- Dy GK, Govindan R, Velcheti V. Long-term outcomes with sotorasib in pretreated KRAS p. G12C-mutated NSCLC: 2-year analysis of CodeBreaK 100. In: AACR Annual Meeting. 2022:abstr CT008.
- Hallin J, Engstrom LD, Hargis L, et al. The KRAS(G12C) Inhibitor MRTX849 Provides Insight toward Therapeutic Susceptibility of KRAS-Mutant Cancers in Mouse Models and Patients. Cancer Discov 2020;10:54-71. [Crossref] [PubMed]
- Sabari JK, Velcheti V, Shimizu K, et al. Activity of Adagrasib (MRTX849) in Brain Metastases: Preclinical Models and Clinical Data from Patients with KRASG12C-Mutant Non-Small Cell Lung Cancer. Clin Cancer Res 2022;28:3318-28. [Crossref] [PubMed]
- Ou SI, Jänne PA, Leal TA, et al. First-in-Human Phase I/IB Dose-Finding Study of Adagrasib (MRTX849) in Patients With Advanced KRAS(G12C) Solid Tumors (KRYSTAL-1). J Clin Oncol 2022;40:2530-8. [Crossref] [PubMed]
- Jänne PA, Riely GJ, Gadgeel SM, et al. Adagrasib in Non-Small-Cell Lung Cancer Harboring a KRAS(G12C) Mutation. N Engl J Med 2022;387:120-31. [Crossref] [PubMed]
- Sacher A, Patel MR, Miller WH, et al. OA03.04 Phase IA Study to Evaluate GDC-6036 Monotherapy in Patients with Non-small Cell Lung Cancer (NSCLC) with KRAS G12C Mutation. J Thorac Oncol 2022;17:S8-9. [Crossref]
- Tanaka N, Lin JJ, Li C, et al. Clinical Acquired Resistance to KRAS(G12C) Inhibition through a Novel KRAS Switch-II Pocket Mutation and Polyclonal Alterations Converging on RAS-MAPK Reactivation. Cancer Discov 2021;11:1913-22. [Crossref] [PubMed]
- Koga T, Suda K, Fujino T, et al. KRAS Secondary Mutations That Confer Acquired Resistance to KRAS G12C Inhibitors, Sotorasib and Adagrasib, and Overcoming Strategies: Insights From In Vitro Experiments. J Thorac Oncol 2021;16:1321-32. [Crossref] [PubMed]
- Awad MM, Liu S, Rybkin II, et al. Acquired Resistance to KRAS(G12C) Inhibition in Cancer. N Engl J Med 2021;384:2382-93. [Crossref] [PubMed]
- Zhao Y, Murciano-Goroff YR, Xue JY, et al. Diverse alterations associated with resistance to KRAS(G12C) inhibition. Nature 2021;599:679-83. [Crossref] [PubMed]
- Adachi Y, Ito K, Hayashi Y, et al. Epithelial-to-Mesenchymal Transition is a Cause of Both Intrinsic and Acquired Resistance to KRAS G12C Inhibitor in KRAS G12C-Mutant Non-Small Cell Lung Cancer. Clin Cancer Res 2020;26:5962-73. [Crossref] [PubMed]
- Nagasaka M, Potugari B, Nguyen A, et al. KRAS Inhibitors- yes but what next? Direct targeting of KRAS- vaccines, adoptive T cell therapy and beyond. Cancer Treat Rev 2021;101:102309. [Crossref] [PubMed]
- Nichols RJ, Cregg J, Schulze CJ, et al. A next generation tri-complex KRASG12C (ON) inhibitor directly targets the active, GTP-bound state of mutant RAS and may overcome resistance to KRASG12C (OFF) inhibition. Cancer Res 2021;81:abstr 1261.
- Hofmann MH, Gerlach D, Misale S, et al. Expanding the Reach of Precision Oncology by Drugging All KRAS Mutants. Cancer Discov 2022;12:924-37. [Crossref] [PubMed]
- Wang X, Allen S, Blake JF, et al. Identification of MRTX1133, a Noncovalent, Potent, and Selective KRAS(G12D) Inhibitor. J Med Chem 2022;65:3123-33. [Crossref] [PubMed]
- Development Programs. Available online: https://investors.modernatx.com/events-and-presentations/Program-Detail/default.aspx (Last accessed 13/11/22).
- Sun C, Hobor S, Bertotti A, et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep 2014;7:86-93. [Crossref] [PubMed]
- Kruspig B, Monteverde T, Neidler S, et al. The ERBB network facilitates KRAS-driven lung tumorigenesis. Sci Transl Med 2018;10:eaao2565. [Crossref] [PubMed]
- Ryan MB, Fece de la Cruz F, Phat S, et al. Vertical Pathway Inhibition Overcomes Adaptive Feedback Resistance to KRAS(G12C) Inhibition. Clin Cancer Res 2020;26:1633-43. [Crossref] [PubMed]
- Misale S, Fatherree JP, Cortez E, et al. KRAS G12C NSCLC Models Are Sensitive to Direct Targeting of KRAS in Combination with PI3K Inhibition. Clin Cancer Res 2019;25:796-807. [Crossref] [PubMed]
- Molina-Arcas M, Moore C, Rana S, et al. Development of combination therapies to maximize the impact of KRAS-G12C inhibitors in lung cancer. Sci Transl Med 2019;11:eaaw7999. [Crossref] [PubMed]
- Xue JY, Zhao Y, Aronowitz J, et al. Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature 2020;577:421-5. [Crossref] [PubMed]
- Solanki HS, Welsh EA, Fang B, et al. Cell Type-specific Adaptive Signaling Responses to KRASG12C Inhibition. Clin Cancer Res 2021;27:2533-48. [Crossref] [PubMed]
- Kerr DL, Haderk F, Bivona TG. Allosteric SHP2 inhibitors in cancer: Targeting the intersection of RAS, resistance, and the immune microenvironment. Curr Opin Chem Biol 2021;62:1-12. [Crossref] [PubMed]
- Ou SI, Koczywas M, Ulahannan S, et al. A12 the SHP2 inhibitor RMC-4630 in patients with KRAS-mutant non-small cell lung cancer: preliminary evaluation of a first-in-man phase 1 clinical trial. J Thorac Oncol 2020;15:S15-6. [Crossref]
- Brana I, Shapiro G, Johnson ML, et al. Initial results from a dose finding study of TNO155, a SHP2 inhibitor, in adults with advanced solid tumors. J Clin Oncol 2021;39:abstr 3005.
- Hillig RC, Sautier B, Schroeder J, et al. Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction. Proc Natl Acad Sci U S A 2019;116:2551-60. [Crossref] [PubMed]
- Johnson M, Gort E, Pant S, et al. A phase I, open-label, dose-escalation trial of BI 1701963 (SOS1:KRAS inhibitor) in patients with KRAS mutated solid tumours: a snapshot analysis. Poster 542. In: European Society for Medical Oncology (ESMO), 2021.
- Tolcher AW, Khan K, Ong M, et al. Antitumor activity in RAS-driven tumors by blocking AKT and MEK. Clin Cancer Res 2015;21:739-48. [Crossref] [PubMed]
- Yohe ME, Gryder BE, Shern JF, et al. MEK inhibition induces MYOG and remodels super-enhancers in RAS-driven rhabdomyosarcoma. Sci Transl Med 2018;10:eaan4470. [Crossref] [PubMed]
- Jänne PA, Shaw AT, Pereira JR, et al. Selumetinib plus docetaxel for KRAS-mutant advanced non-small-cell lung cancer: a randomised, multicentre, placebo-controlled, phase 2 study. Lancet Oncol 2013;14:38-47. [Crossref] [PubMed]
- Yen I, Shanahan F, Merchant M, et al. Pharmacological Induction of RAS-GTP Confers RAF Inhibitor Sensitivity in KRAS Mutant Tumors. Cancer Cell 2018;34:611-25.e7. [Crossref] [PubMed]
- Capelletto E, Bironzo P, Denis L, et al. Single agent VS-6766 or VS-6766 plus defactinib in KRAS-mutant non-small-cell lung cancer: the RAMP-202 phase II trial. Future Oncol 2022;18:1907-15. [Crossref] [PubMed]
- Sulahian R, Kwon JJ, Walsh KH, et al. Synthetic Lethal Interaction of SHOC2 Depletion with MEK Inhibition in RAS-Driven Cancers. Cell Rep 2019;29:118-34.e8. [Crossref] [PubMed]
- Jones GG, Del Río IB, Sari S, et al. SHOC2 phosphatase-dependent RAF dimerization mediates resistance to MEK inhibition in RAS-mutant cancers. Nat Commun 2019;10:2532. [Crossref] [PubMed]
- Boned Del Río I, Young LC, Sari S, et al. SHOC2 complex-driven RAF dimerization selectively contributes to ERK pathway dynamics. Proc Natl Acad Sci U S A 2019;116:13330-9. [Crossref] [PubMed]
- Terai H, Hamamoto J, Emoto K, et al. SHOC2 Is a Critical Modulator of Sensitivity to EGFR-TKIs in Non-Small Cell Lung Cancer Cells. Mol Cancer Res 2021;19:317-28. [Crossref] [PubMed]
- Xiao-Pei H, Ji-Kuai C, Xue W, et al. Systematic identification of Celastrol-binding proteins reveals that Shoc2 is inhibited by Celastrol. Biosci Rep 2018;38:BSR20181233. [Crossref] [PubMed]
- Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017;168:960-76. [Crossref] [PubMed]
- Örd D, Örd T, Biene T, et al. TRIB3 increases cell resistance to arsenite toxicity by limiting the expression of the glutathione-degrading enzyme CHAC1. Biochim Biophys Acta 2016;1863:2668-80. [Crossref] [PubMed]
- Erazo T, Lorente M, López-Plana A, et al. The New Antitumor Drug ABTL0812 Inhibits the Akt/mTORC1 Axis by Upregulating Tribbles-3 Pseudokinase. Clin Cancer Res 2016;22:2508-19. [Crossref] [PubMed]
- Roh JI, Lee J, Sung YH, et al. Impaired AKT signaling and lung tumorigenesis by PIERCE1 ablation in KRAS-mutant non-small cell lung cancer. Oncogene 2020;39:5876-87. [Crossref] [PubMed]
- Bressler KR, Ross JA, Ilnytskyy S, et al. Depletion of eukaryotic initiation factor 5B (eIF5B) reprograms the cellular transcriptome and leads to activation of endoplasmic reticulum (ER) stress and c-Jun N-terminal kinase (JNK). Cell Stress Chaperones 2021;26:253-64. [Crossref] [PubMed]
- Moll HP, Pranz K, Musteanu M, et al. Afatinib restrains K-RAS-driven lung tumorigenesis. Sci Transl Med 2018;10:eaao2301. [Crossref] [PubMed]
- Pastushenko I, Mauri F, Song Y, et al. Fat1 deletion promotes hybrid EMT state, tumour stemness and metastasis. Nature 2021;589:448-55. [Crossref] [PubMed]
- Kitai H, Ebi H, Tomida S, et al. Epithelial-to-Mesenchymal Transition Defines Feedback Activation of Receptor Tyrosine Kinase Signaling Induced by MEK Inhibition in KRAS-Mutant Lung Cancer. Cancer Discov 2016;6:754-69. [Crossref] [PubMed]
- Li BT, Falchook GS, Durm GA, et al. OA03. 06 codebreak 100/101: First report of safety/efficacy of sotorasib in combination with pembrolizumab or atezolizumab in advanced KRAS p. G12C NSCLC. J Thorac Oncol 2022;17:S10-1. [Crossref]


