
CKAP4 is a potential exosomal biomarker and therapeutic target for lung cancer
Highlight box
Key findings
• CKAP4 was detectable in the sera of lung cancer patients using ELISA, and serum CKAP4 levels reflected tumor expression of CKAP4.
• An anti-CKAP4 antibody in combination with osimertinib showed synergistic anti-tumor effect against HCC4006 cells.
What is known and what is new?
• The binding of DKK1 to CKAP4 promotes tumor cell growth and simultaneous expression of both proteins are correlated with poor prognosis in pancreatic, esophageal, and lung cancer.
• After binding with DKK1, CKAP4 is secreted with exosome in pancreatic cancer, which is detectable by ELISA.
• An anti-CKAP4 antibody shows anti-tumor effects via inhibition of DKK1 binding to CKAP4.
• CKAP4 is modified with palmitic acid, which is required for promoting tumor cell growth.
What is the implication, and what should change now?
• A combined anti-CKAP4 antibody and osimertinib may be a novel therapeutic strategy for lung cancer.
• Serum CKAP4 is a biomarker for lung cancer, and it may be a companion diagnostic for anti-CKAP4 antibody.
• CKAP4 secretion with exosomes are regulated by palmitoylation.
Introduction
Worldwide, lung cancer is the leading cause of cancer-related death among men and women (1). Approximately 85% of lung cancers are non-small cell lung cancer (NSCLC) consisting of adeno-, squamous-, and large-cell undifferentiated carcinomas (2). Of these, adenocarcinoma is the most common histological subtype, and over half of patients with NSCLC presenting advanced stages and receiving platinum-based chemotherapy. In recent decades, different drug classes, including molecular targeted therapies, such as epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) and immune checkpoint inhibitors, have improved outcomes for patients with advanced NSCLC (3). In spite of diagnostic and treatment advances, NSCLC prognoses remain poor, therefore developing new molecular targeted drugs for lung cancer, where TKIs and immune checkpoint inhibitors do not work, are required.
Cytoskeleton-associated protein 4 (CKAP4; also known as CLIMP-63 and ERGIC-63) was originally identified as a non-glycosylated type II transmembrane protein in the endoplasmic reticulum (ER) (4,5) with several functions (6-10). In addition to ER localization, CKAP4 was identified in the plasma membrane (PM) and functioned as a receptor of the extracellular protein Dickkopf-1 (DKK1) (11,12). Clinical evidence suggested that DKK1 is positively involved in tumorigenesis, but the underlying mechanism where DKK1 promotes tumor growth has remained unclear for a long time (13-15). It has been found that the binding of DKK1 to CKAP4 activates Ak strain transforming (AKT) via phosphoinositide 3-kinase (PI3K) and that overexpression of DKK1 and CKAP4 is observed in pancreatic, esophageal, and liver cancers (11,16-18), while an anti-CKAP4 antibody, which blocks DKK1 binding to CKAP4, inhibited in vivo tumor formation by pancreatic cancer cells (19). In terms of lung cancer, immunohistochemically high DKK1 expression levels in NSCLC patients are associated with poor prognoses, and serum DKK1 levels have been shown to become a clinical marker (20,21). CKAP4 is also highly immunohistochemically expressed in NSCLC patients (11).
Reverse-phase protein array studies reported that serum CKAP4 levels in lung cancer patients are higher than those of healthy controls (HCs) (22), suggesting that CKAP4 may be a novel early sero-diagnostic marker for lung cancer. Indeed, CKAP4 was released with exosomes in the extracellular space via the biogenesis pathway, from the PM to multivesicular bodies (MVBs) via early endosomes, and was detected using enzyme-linked immunosorbent assay (ELISA) in sera from pancreatic cancer patients (19). In addition, CKAP4 was modified with palmitate at Cys100 by palmitoyl acyltransferase DHHC2 (23,24), and palmitoylation was required for CKAP4 localization to PM lipid rafts to promote cancer cell proliferation (25). Palmitoylation was also involved in the formation of a complex between CKAP4 and voltage-dependent anion-selective channel protein 2, and for mitochondrial membrane potential maintenance (10). However, it is unclear if palmitoylation affects CKAP4 secretion with exosomes.
Extending our previous observations, we asked whether anti-CKAP4 antibody is available for the diagnosis and treatment of lung cancer. The current study aimed to show not only the simple application of the findings in pancreatic cancer to lung cancer but also the novel findings about the biogenesis and trafficking of exosomes with CKAP4 and the combination of anti-CKAP4 antibody and standard therapy. Here we show that CKAP4 is detected at higher rates in sera from NSCLC patients, and that palmitoylation promotes CKAP4 secretion with exosomes. Additionally, combination treatments with an anti-CKAP4 antibody and osimertinib, a third generation EGFR-TKI, strongly inhibited in vivo tumor formation by lung cancer cells compared with monotherapy. We present the following article in accordance with the ARRIVE reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-571/rc).
Methods
Materials and chemicals
All antibodies, chemicals, and cell lines are shown in Tables S1-S3. A549, Calu-1, NCI-H292, and HCC4006 cells were authenticated in July 2022 using short tandem repeat analysis and mycoplasma testing (MycoStrip, InvivoGen). Target sequences for siRNA and shRNA assays are shown in Table S4. Primer sequences for quantitative polymerase chain reaction (PCR) are shown in Table S5. Anti-CKAP4 monoclonal antibodies were generated as previously described (19). Knockout (KO) cells were generated using the CRISPR/Cas9 system as previously described (26).
CKAP4WT-HA and CKAP4C100S-HA are wild-type (WT) CKAP4 and CKAP4 mutant in which cysteine at amino acid number 100 was changed to serine, respectively. CKAP4C100S-HA is not palmitoylated. Both CKAP4s were tagged by HA at the C-terminal end.
Human serum and tissues samples
Tissue samples were obtained from 92 NSCLC patients (median age =69; range, 40–92 years) who underwent surgical resection at Osaka University Hospital (Osaka, Japan) between June 2018 and August 2019. NSCLC patients were newly presented and previously untreated, and their diagnoses were pathologically confirmed. Serum samples from age- and sex-matched 92 HCs were used as controls and came from pooled HC serum samples at Osaka University Hospital. In NSCLC patients, no cancerous lesions were detected other than in the lung. In HCs, no symptoms or laboratory findings suggested a cancer history. In eight NSCLC cases, pre- and post-operative sera were obtained. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the ethics review board of the Graduate School of Medicine, Osaka University, Japan (Nos. 10026, 13455, 13563-8, and 18528-8) and informed consent was provided by all the patients.
Histology
Surgically resected specimens were fixed in 10% formalin and paraffin embedded. Specimens were stored at room temperature (RT) in the dark, sectioned at 4 µm, and subjected to CKAP4 and DKK1 immunohistochemistry (IHC) analysis. In surgical resection specimens, tumors where positive CKAP4 and DKK1 staining covered <5%, 5–20% and 20–50%, and >50% of the total area were classified as CKAP4- or DKK1-negative, low, moderate, and high expression, respectively. At least three investigators evaluated the sections independently in a blinded fashion.
Statistical analyses
All experiments were repeated at least three times, and results were expressed as the mean ± standard deviation (SD) or standard error (SE). Statistical analyses were performed using JMP version 16 and SAS version 9.4 (SAS Institute Inc., Cary, NC, USA). The mean and median of continuous outcome variables were tested using Student’s t-test and Mann-Whitney U test, respectively. Cumulative probability values of overall survival and relapse-free survival rates were calculated using the Kaplan-Meier method, and generalized Wilcoxon tests were used to assess their statistical significance. P values <0.05 were considered statistically significant. Western blotting data were representative of at least three independent experiments.
Sandwich enzyme-linked immunosorbent assay (ELISA) for assaying CKAP4 concentration in small extracellular vesicles (SEVs)
A sandwich ELISA for measuring CKAP4 concentrations in serum was performed as previously described, with modifications (19). Briefly, 96-well plates were coated with 80 µL of anti-CKAP4 antibody (3F11-2B10; 2 µg/mL) and incubated overnight at RT. Wells were washed and then blocked for 1 h at RT. After washing, 80 µL of undiluted serum sample was added to wells and incubated for 2 h at RT. After washing, 80 µL of biotinylated anti-CKAP4 antibody (1G4-4A9; 0.1 µg/mL) was added to wells and incubated for 2 h at RT. After washing, 80 µL of horseradish peroxidase (HRP)-streptavidin (DY998, R&D Systems, Inc.) was added to wells and incubated for 20 min at RT. After washing, a substrate solution (DY999, R&D Systems, Inc.) was added to wells, reacted for 30 min, and stopped by adding 80 µL of STOP solution (Cell Signaling Technology). Color intensity was calculated by subtracting optical density readings at 540 nm from readings at 450 nm on a multimode plate reader. A standard reference curve was prepared using recombinant CKAP4 protein serial dilutions. When the CKAP4 concentration was <0.1 ng/mL (the lower limit of the standard curve), it was defined as zero.
Exosome isolation
A549 cells (5×106 cells) cultured in 10-cm culture dish were washed with PBS buffer for three times, and then serum free Dulbecco’s modified Eagle’s medium (DMEM) was added, and cells were cultured for 2 days. The cultured medium was collected and passed 0.22 µm syringe filter and stocked as conditioned medium (CM). To isolate and purify exosomes from serum samples or CM, sequential centrifugation or affinity precipitation (AP) methods were conducted as previously described (19). For sequential centrifugation, 50 mL of CM from cultured cells was subjected to sequential centrifugation: 2,000 ×g for 20 min, 10,000 ×g for 30 min, and 100,000 ×g for 70 min. Ultracentrifugation was performed using a SW55Ti swinging bucket rotor (Beckman Coulter, Brea, CA, USA). SEVs were then washed in phosphate buffered saline (PBS) and precipitated by ultracentrifugation at 100,000 ×g for 30 min. In AP methods, exosomes were precipitated from 100 mL of CM using the MagCaptureTM Exosome Isolation Kit PS (Wako, Fujifilm Wako Pure Chemical Corporation, Japan) according to manufacturer’s recommendations.
Purification of lysosomal proteins
To isolate and purify lysosomal proteins from cultured cells, magnetic activated cell sorting system (MACS) using dextran-coated ferrofluid (DexoMAG 40, Liquids Research Ltd., Gwynedd, Wales, UK) was performed based on manufacturer’s instructions, with some modifications. Briefly, A549/CKAP4 KO/CKAP4 WT-HA or A549/CKAP4 KO/CKAP4C100S-HA cells (1×107 cells) were plated onto biotinylated surfaces in 100-mm culture dishes and cultured for 24 h in DMEM supplemented with 10% fetal calf serum containing (FCS) 10% (v/v) DexoMAG and 10 nM bafilomycin so that magnetic particles were endocytosed and captured in the endocytotic pathway. Next, cells were washed three times in PBS to remove free DexoMAG, and cultured for another 24 h in serum-starved culture conditions (bafilomycin treatment was continued). Cells were then mechanically collected and suspended in 2 mL of ice cold buffer A [10 mM HEPES-NaOH (pH 7.0), 15 mM KCl, 1.5 mM MgAc, and 1 mM dithiothreitol (DTT)] supplemented with protease inhibitors [10 µg/mL leupeptin, 10 µg/mL aprotinin, and 50 µg/mL phenylmethylsulfonyl fluoride (PMSF)]. Cells were then homogenized by passing through a 26- and 30-gauge needle (10 times for both), followed by Potter-Elehjem homogenization (60 strokes). Cell lysates were mixed with 500 µL of buffer B [220 mM HEPES-NaOH (pH 7.0), 0.1 mM sucrose, 375 mM KCl, 225 mM MgAc, and 1 mM DTT] and aliquots were centrifuged at 150 ×g for 10 min to yield nuclear pellets. Supernatants containing subcellular components, including the lysosomes, early endosomes, and late endosomes were separated using the MiniMACS Separator (Miltenyi Biotec). MS columns were blocked and equilibrated in blocking buffer (PBS plus 0.5% bovine serum albumin) and supernatants were loaded (subcellular components containing DexoMAG were magnetically captured in the MS column). After column washing three times in washing buffer (PBS plus 0.1 mM sucrose), the MS column was detached from the magnetic stand and lysosomal- and other endosomal-components were eluted and collected in elution buffer (PBS containing 0.1 mM sucrose and protease inhibitors). To isolate endocytosed CKAP4 proteins from the total lysosomal CKAP4 proteins, CKAP4 proteins located on the PM were biotinylated with EZ-LinkTM Sulfo-NHS-Biotin (Thermo Scientific, Waltham, MA, USA) before DexoMAG treatment. After MACS purification of total lysosomal proteins, Noidet-P40 was added to the eluate (final concentration =1%) and biotinylated proteins (lysosomal proteins endocytosed from the PM) were affinity precipitated with 30 µL streptavidin magnetic beads (Invitrogen, CA, USA). After bead washing, precipitates were eluted in sample buffer.
Discontinuous sucrose gradient
Density gradients centrifugation were performed to purify SEVs. Isolated SEVs were suspended in 2.5 M sucrose. The SEV suspension was overlaid with a step sucrose gradient (2.0–0.25 M sucrose) and was ultracentrifuged at 100,000 ×g for 2.5 h in a Beckman SW41Ti rotor (19). Fractions (1 mL each) were collected from top to bottom and SEVs were concentrated by centrifugation at 100,000 ×g for 70 min.
Three-dimension (3D) cell proliferation assays
3D cell proliferation assays were performed as previously described, with some modifications (11,27,28). To analyze sphere growth, cells were seeded in Matrigel (BD Biosciences, San Jose, CA, USA); 40 µL of Matrigel was added to a round coverslip and solidified for 30 min at 37 ℃. Cells (1×104) suspended in DMEM plus 10% FCS and 2% Matrigel were added onto the solidified Matrigel and cultured for 5 days.
Plasmid construction and lentivirus production
Lentivirus vectors were constructed by inserting relevant cDNAs into CSII-CMV-MCS-IRES2-Bsd plasmids kindly provided by Dr. H. Miyoshi (RIKEN BioResource Center, Ibaraki, Japan) or the pLVSIN-CMV Puro vector (Takara Bio Inc., Shiga, Japan). To construct lentivirus vectors harboring shRNAs, an oligonucleotide DNA fragment containing the H1 promoter and shRNA were cloned into the CS-RfA-EVBsd vector using Gateway technology (Invitrogen) or MISSION TRC shRNAs (shDKK1#1: TRCN0000033385, Sigma Aldrich, St. Louis, MO, USA). Lentivirus particles were generated by transfection of those plasmids harboring cDNAs or shRNAs concomitant with pCAG-HIV-gp and pCMV-VSV-G-RSV-Rev into X293T cells using FuGENE HD transfection reagent (Promega).
Xenograft tumor assay
Xenograft tumor assays were performed as previously described, with some modifications (27). Six-week-old male BALB/cAJcl-nu/nu immunodeficient mice (CLEA, Tokyo, Japan) were anesthetized with a combination of medetomidine (0.3 mg/kg body weight), midazolam (4 mg/kg), and butorphanol (5.0 mg/kg). Mice then received a dorsal subcutaneous transplantation of NCI-H292 or HCC4006 cells (5×106 cells, respectively) in 200 µL of PBS, and tumor volumes were allowed to reach 50 mm3. Then, an anti-CKAP4 antibody (3F11-2B10, 200 µg) or isotype control IgG were intraperitoneally injected twice a week. In the combination therapy model, mice were daily orally gavaged with osimertinib (1.25 mg/kg body weight) or a vehicle control. Tumor development continued for an additional 19 days (NCI-H292 cells) or 28 days (HCC4006 cells), after which mice were humanely sacrificed. Resected tumors were measured and weighed. Tumor volumes were calculated using the following formula: (length) × (width) × (height)/2. Xenograft tumor volumes <50 mm3 at treatment initiation were excluded from analyses. Mice were housed in a standardized environment with good ventilation, 12:12 h light/dark cycle, and with free access to food pellets and water. No randomization was used to allocate mice to cell lines and no blinding was performed. All animal experiments in this study were performed under a project license (No. 21-048-1) granted by the Animal Research Committee of Osaka University, Japan, in compliance with the Guidelines for Animal Experiments of Osaka University for the care and use of animals. A protocol was prepared before the study without registration. Sample sizes were empirically chosen based on optimizing the maximum number of samples per independent experiment while considering the 3Rs (reduction/refinement/replacement) of animal research.
Acyl-PEGyl exchange gel shift (APEGS) assay
The APEGS assay was performed as previously described, with some modifications (25). A549, A549/CKAP4 KO/CKAP4-HA, and A549/CKAP4 KO/CKAP4C100S-HA cells (subconfluent in 100-mm culture dishes) were cultured in serum free DMEM for 24 h. Cells were rinced with ice cold PBS containing 1 mM CaCl2 and MgCl2 three times, and then incubated with EZ-LinkTM Sulfo-NHS-biotin (0.5 mg/mL) for 30 min at 4 ℃. Free S-NHS-biotin was quenched by washing cells three times in ice-cold PBS plus 50 mM NH4Cl and then washing once in ice-cold PBS buffer. Cells were lysed in 1 mL of lysis buffer [PBS containing 5 mM ethylenediaminetetraacetic acid (EDTA) and 4% sodium dodecyl sulfate (SDS)] supplemented with protease inhibitors. After sonication and centrifugation at 20,000 ×g for 15 min at RT, proteins (0.4–0.5 mg/mL in 1 mL) were reduced in 25 mM tris(2-carboxyethyl)phosphine (TCEP) for 1 h at 55 ℃, and free cysteine residues were alkylated in 50 mM N-ethylmaleimide for 3 h at RT. After chloroform-methanol precipitation (CM ppt), pellets were resuspended in 250 µL of lysis buffer supplemented with pepstatin A (10 µg/mL), and then centrifuged at 20,000 ×g for 10 min at RT to completely remove undissolved protein pellets. Supernatants (125 µL) were mixed with 375 µL of hydroxylamine (HAM) buffer [1.33 M hydroxylamine (pH 7.0) containing 0.2% Triton X-100, and 5 mM EDTA] or control buffer [1.33 M Tris-HCl (pH 7.0), 0.2% Triton X-100, and 5 mM EDTA], and mixtures were incubated for 1 h at 37 ℃. After the CM ppt, pellets were resuspended in 150 µL of lysis buffer supplemented with pepstatin (10 µg/mL). Soluble proteins (0.5–0.75 mg/mL in 100 µL) were PEGylated with 20 mM mPEG-5k in the presence of 10 mM TCEP for 1 h so that newly exposed cysteinyl-thiol groups were labeled with mPEG-5k. After CM ppt, pellets were resuspended in 500 µL of resuspension buffer [25 mM HEPES-NaOH (pH 7.4) containing 150 mM NaCl, 5 mM EDTA, 0.1% SDS, and 1% Triton X-100] and centrifuged at 20,000 ×g for 10 min at 4 ℃. PM proteins in supernatants were pulled down by NeutrAvidin agarose beads, and eluates were probed with indicated antibodies. Protein concentrations were measured using the bicinchoninic acid protein assay at different steps.
Others
The cell proliferation assay was performed as described (n=3) (25). A549/CKAP4 KO cells were generated by CRISPR/Cas9 as previously described (26). Control A549 and Calu-1 cells in the experiments using short-hairpin RNA (shRNA) were generated by transfection with the empty viral vector. To generate A549 and NCI-H292 cells stably expressing proteins, parental cells (5×104 cells/well in 12-well plates) were transfected with the lentivirus and selected using blasticidin S or puromycin. Knockdown of protein expression by siRNA and quantitative PCR were performed as previously described (19,27,28). For Western blotting, clathrin and HSP90 were used as loading controls.
Results
CKAP4 is secreted from lung cancer cells with exosomes
CKAP4 was expressed in different cells, including non-tumor and cancer cells (14), but its PM localization was variable. DKK1 and CKAP4 expression levels were examined in eight NSCLC cell lines (Figure 1A). CKAP4 was present in the PMs of A549, Calu-1, NCI-H1573, NCI-H1650, and NCI-358 cells; however, it was barely detectable in the PMs of NCI-H1975, NCI-H292, or NCI-H441 cells (Figure 1A). DKK1 was only expressed in A549, Calu-1, and NCI-H292 cells (Figure 1A). CKAP4 was detected in SEVs including exosomes, which were isolated from the CM of A549 and Calu-1 cells using Tim4 (29), a phosphatidylserine-binding protein that recognizes exosomes, whereas CKAP4 was present at lower levels in exosomes from other cells (Figure 1A). CKAP4 localization in exosomes was further confirmed using a discontinuous sucrose gradient. CKAP4 from A549 cell CM was sedimented with exosome marker proteins (CD81 and TSG101) at a density of 1.130–1.168 g/mL (Figure 1B). DKK1 induces clathrin-mediated CKAP4 endocytosis to enhance its secretion with exosomes (19). CKAP4-containing exosomes in 100,000 ×g CM precipitates were decreased by DKK1 knockdown, but rescued by DKK1 expression (Figure 1C). This observation was also confirmed using Tim4 (Figure S1A).
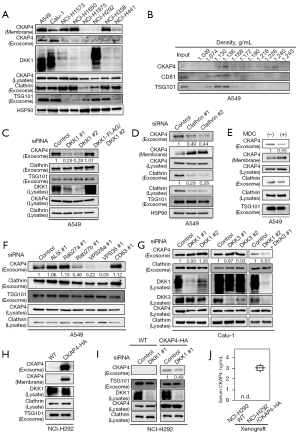
Clathrin knockdown or monodansyl cadaverine (MDC) treatment, which impairs clathrin-mediated endocytosis (30), inhibited CKAP4 secretion with exosomes from A549 cells; PM-localized CKAP4 levels increased in reverse proportion to CKAP4 levels in exosomes (Figure 1D,1E). Consistent with the notion that exosomes were derived from the endosomal origin (31), exosomal CKAP4 is likely to primarily locate to the PM and reflects cell surface CKAP4 expression in lung cancer cells.
Many proteins are involved in exosome biogenesis (31), inducing endosomal sorting complexes required for transport and associated proteins (TSG101, HRS, and ALIX), tetraspanin proteins (CD9, CD63, and CD81), small G proteins (Rab27a and Rab27b), and proteins involved in vacuole protein sorting (VPS) (VPS26a and VPS35). The protein composition of exosomes varies by vesicle (32). When exosome markers were depleted in A549 cells, CKAP4 in SEVs was clearly decreased in Rab27b-, VPS26a-, and VPS35-depleted cells (Figure 1F, Figure S1B,S1C).
DKK1 knockdown in Calu-1 cells did not affect exosomal CKAP4 (Figure 1G). DKK3, which functions as another ligand for CKAP4 (16), was also expressed in Calu-1 cells. Exosomal CKAP4 levels decreased in DKK1 and DKK3 double-depleted cells (Figure 1G). NCI-H292 cells did not secrete CKAP4 with exosomes because CKAP4 was barely expressed in the cells (Figure 1A). When CKAP4-HA was exogenously expressed in NCI-H292 cells (NCI-H292/CKAP4-HA), CKAP4 was detected in the PM (Figure 1H). Additionally, CKAP4 was detected in exosomes secreted from NCI-H292/CKAP4-HA cells, while DKK1 knockdown decreased these levels (Figure 1I). Control NCI-H292 or NCI-H292/CKAP4-HA cells were subcutaneously injected into immuno-deficient mice. Mice were bled for three weeks and CKAP4 was quantitatively detected in the serum of mice injected with NCI-H292/CKAP4-HA cells using sandwich ELISA, whereas serum CKAP4 was not detected in mice transplanted with WT NCI-H292 cells (Figure 1J). Thus, CKAP4 was secreted with exosomes from lung cancer cells where DKK1 is expressed and CKAP4 is in the PM.
CKAP4 is detected in NSCLC patient sera
To examine the clinical relevance of CKAP4 expression in NSCLC, DKK1 and CKAP4 IHC analyses were performed in 92 NSCLC patients (Tables 1-3). Stained areas were classified into four categories (<5%, 5–20%, 20–50%, and >50%), which corresponded to negative, low, moderate, and high expression, respectively (Figure 2A, Figure S2). Staining for both proteins was considered positive when stained areas had more than low expression. Based on these criteria, CKAP4 and DKK1 were positive for 76% and 44% of the 92 cases, respectively. Both proteins were minimally detected in non-tumor regions (Figure 2A). When patients with positive and negative expression for either DKK1 or CKAP4 were compared, no significant difference was observed with respect to clinicopathological parameters (Tables 1,2). However, when patients positive for both proteins were compared with other patients, T grades (tumor size and invasion depth) were significantly higher in the former group (Table 3). Consistent with these results, patients positive for both proteins were associated with worse relapse-free survival (P=0.011) and overall survival (P=0.028) rates when compared with other patients (Figure 2B,2C).
Table 1
| Parameters | CKAP4 positive (n=70) | CKAP4 negative (n=22) | P value |
|---|---|---|---|
| Age (years) | 70 (40–92) | 70 (48–83) | 0.621 |
| Sex (male/female) | 47/23 | 13/9 | 0.489 |
| Preoperative CEA (ng/mL) | 3 (0–51) | 3 (0–30) | 0.856 |
| Histology (Ad/Sq/others) | 55/12/3 | 14/7/1 | 0.325 |
| pT (Tis-2/3, 4) | 59/11 | 20/2 | 0.437 |
| pN (0–1/2) | 68/2 | 20/2 | 0.211 |
| Lymphatic vessel invasion (ly0/1) | 56/14 | 17/5 | 0.783 |
| Venous invasion (v0/1) | 65/5 | 21/1 | 0.667 |
| Pleural invasion (pl0/1–3) | 65/5 | 21/1 | 0.667 |
| pStage (0–II/III, IV) | 59/11 | 18/4 | 0.785 |
Continuous values are shown as the median (range), whereas categorical values are shown as the total number. Tis, carcinoma in situ. T1, tumor greatest dimension is ≤3 cm, surrounded by lung or visceral pleura, without bronchoscopic evidence of invasion more proximal than the lobar bronchus. T2, tumor >3 cm but ≤5 cm or with any of the following features: involves main bronchus, invades visceral pleura, or is associated with atelectasis or obstructive pneumonitis that extends to the hilar region but does not involve the entire lung. T3, tumor >5 cm but ≤7 cm or with any of the following features: directly invades parietal pleura, chest wall (including superior sulcus tumors), phrenic nerve, or pericardium; or separate tumor nodule(s) in the same lobe. T4, tumor >7 cm or one that invades any of the following: diaphragm, mediastinum, heart, great vessels, trachea, recurrent laryngeal nerve, esophagus, spine or carina; or separate tumor nodule(s) in a different ipsilateral lobe. N0, no regional lymph node metastasis. N1, metastasis in ipsilateral, peribronchial, and/or ipsilateral hilar lymph nodes and intrapulmonary nodes, including involvement by direct extension. N2, metastasis in ipsilateral mediastinal and/or subcarinal lymph node(s). ly0, no lymphatic vessel invasion. ly1, lymphatic vessel invasion. v0, no venous invasion. v1, venous invasion. pl0, no pleural invasion. pl1, tumor histologically invade the external elastic layer of visceral pleura but does not reach outer surface of the visceral pleura. pl2, tumor is histologically exposed on the pleural surface. pl3, tumor histologically invades any of the following: chest wall, diaphragm, mediastinum, or adjacent lobe. CKAP4, cytoskeleton-associated membrane protein 4; CEA, carcinoembryonic antigen; Ad, adenocarcinoma; Sq, squamous cell carcinoma.
Table 2
| Parameters | DKK1 positive (n=51) | DKK1 negative (n=41) | P value |
|---|---|---|---|
| Age (years) | 70 (40–92) | 70 (48–82) | 0.832 |
| Sex (male/female) | 33/18 | 27/14 | 0.909 |
| Preoperative CEA (ng/mL) | 3 (0–30) | 3 (0–51) | 0.922 |
| Histology (Ad/Sq/others) | 42/7/2 | 27/12/2 | 0.171 |
| pT (Tis-2/3, 4) | 41/10 | 38/3 | 0.093 |
| pN (0–1/2) | 50/1 | 38/3 | 0.211 |
| Lymphatic vessel invasion (ly0/1) | 41/10 | 32/9 | 0.783 |
| Venous invasion (v0/1) | 48/3 | 38/3 | 0.782 |
| Pleural invasion (pl0/1–3) | 47/4 | 39/2 | 0.567 |
| pStage (0–II/III, IV) | 44/7 | 33/8 | 0.455 |
Continuous values are shown as the median (range), whereas categorical values are shown as the total number. Tis, carcinoma in situ. T1, tumor greatest dimension is ≤3 cm, surrounded by lung or visceral pleura, without bronchoscopic evidence of invasion more proximal than the lobar bronchus. T2, tumor >3 cm but ≤5 cm or with any of the following features: involves main bronchus, invades visceral pleura, or is associated with atelectasis or obstructive pneumonitis that extends to the hilar region but does not involve the entire lung. T3, tumor >5 cm but ≤7 cm or with any of the following features: directly invades parietal pleura, chest wall (including superior sulcus tumors), phrenic nerve, or pericardium; or separate tumor nodule(s) in the same lobe. T4, tumor >7 cm or one that invades any of the following: diaphragm, mediastinum, heart, great vessels, trachea, recurrent laryngeal nerve, esophagus, spine or carina; or separate tumor nodule(s) in a different ipsilateral lobe. N0, no regional lymph node metastasis. N1, metastasis in ipsilateral, peribronchial, and/or ipsilateral hilar lymph nodes and intrapulmonary nodes, including involvement by direct extension. N2, metastasis in ipsilateral mediastinal and/or subcarinal lymph node(s). ly0, no lymphatic vessel invasion. ly1, lymphatic vessel invasion. v0, no venous invasion. v1, venous invasion. pl0, no pleural invasion. pl1, tumor histologically invade the external elastic layer of visceral pleura but does not reach outer surface of the visceral pleura. pl2, tumor is histologically exposed on the pleural surface. pl3, tumor histologically invades any of the following: chest wall, diaphragm, mediastinum, or adjacent lobe. DKK1, Dickkopf-1; CEA, carcinoembryonic antigen; Ad, adenocarcinoma; Sq, squamous cell carcinoma.
Table 3
| Parameters | CKAP4 (+) and DKK1 (+) (n=42) | CKAP4 (−) and/or DKK1 (−) (n=50) | P value |
|---|---|---|---|
| Age (years) | 70 (40–92) | 70 (48–83) | 0.922 |
| Sex (male/female) | 29/13 | 31/19 | 0.480 |
| Preoperative CEA (ng/mL) | 3 (0–24) | 3 (0–51) | 0.760 |
| Histology (Ad/Sq/others) | 35/5/2 | 34/14/2 | 0.165 |
| pT (Tis-2/3, 4) | 32/10 | 47/3 | 0.015 |
| pN (0–1/2) | 41/1 | 47/3 | 0.397 |
| Lymphatic vessel invasion (ly0/1) | 33/9 | 40/10 | 0.866 |
| Venous invasion (v0/1) | 39/3 | 47/3 | 0.825 |
| Pleural invasion (pl0/1–3) | 38/4 | 48/2 | 0.285 |
| pStage (0–II/III, IV) | 35/7 | 42/8 | 0.931 |
Continuous values are shown as the median (range), whereas categorical values are shown as the total number. Tis, carcinoma in situ. T1, tumor greatest dimension is ≤3 cm, surrounded by lung or visceral pleura, without bronchoscopic evidence of invasion more proximal than the lobar bronchus. T2, tumor >3 cm but ≤5 cm or with any of the following features: involves main bronchus, invades visceral pleura, or is associated with atelectasis or obstructive pneumonitis that extends to the hilar region but does not involve the entire lung. T3, tumor >5 cm but ≤7 cm or with any of the following features: directly invades parietal pleura, chest wall (including superior sulcus tumors), phrenic nerve, or pericardium; or separate tumor nodule(s) in the same lobe. T4, tumor >7 cm or one that invades any of the following: diaphragm, mediastinum, heart, great vessels, trachea, recurrent laryngeal nerve, esophagus, spine or carina; or separate tumor nodule(s) in a different ipsilateral lobe. N0, no regional lymph node metastasis. N1, metastasis in ipsilateral, peribronchial, and/or ipsilateral hilar lymph nodes and intrapulmonary nodes, including involvement by direct extension. N2, metastasis in ipsilateral mediastinal and/or subcarinal lymph node(s). ly0, no lymphatic vessel invasion. ly1, lymphatic vessel invasion. v0, no venous invasion. v1, venous invasion. pl0, no pleural invasion. pl1, tumor histologically invade the external elastic layer of visceral pleura but does not reach outer surface of the visceral pleura. pl2, tumor is histologically exposed on the pleural surface. pl3, tumor histologically invades any of the following: chest wall, diaphragm, mediastinum, or adjacent lobe. DKK1, Dickkopf-1; CKAP4, cytoskeleton-associated membrane protein 4; CEA, carcinoembryonic antigen; Ad, adenocarcinoma; Sq, squamous cell carcinoma.

Serum CKAP4 levels were also measured in 92 lung cancer patients and matched HCs. When CKAP4 values >0.1 ng/mL were considered positive, 18 cases were positive in NSCLC patients (19.6%) and six cases were positive in HCs (6.5%) (Figure 2D). Of note, high serum CKAP4 values (>10 ng/mL) were only observed in NSCLC patients. Patients with IHC-positive CKAP4 levels had significantly higher CKAP4 serum positive rates (17/70, 24.3%) when compared with patients with negative CKAP4 IHC values (1/22, 4.5%) (Figure 2E). Postoperative sera were obtained from eight patients whose preoperative serum CKAP4 values were positive. In all these samples, postoperative CKAP4 levels were reduced (Figure 2F). Taken together, in our cohort, serum CKAP4 levels reflected CKAP4 expression in lung cancer lesions.
CKAP4 palmitoylation enhances CKAP4 secretion with exosomes
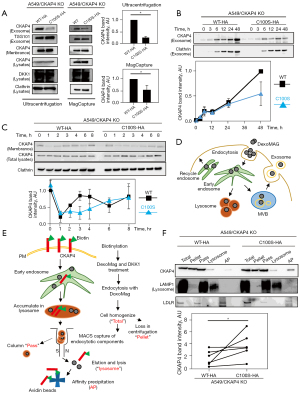
CKAP4 is palmitoylated at Cys100, which is required for CKAP4 localization to lipid rafts to activate DKK1-CKAP4 signaling (25). To examine the role of palmitoylation in conjunction with CKAP4 secretion with exosomes, WT-CKAP4 or CKAP4C100S, which could not be modified with palmitate (Figure S3A), were expressed in CKAP4 KO A549 cells (A549/CKAP4 KO/CKAP4-HA and A549/CKAP4 KO/CKAP4C100S-HA cells). Although basal PM-CKAP4 levels were similar between the two cells (Figure 3A, Membrane), exosomal CKAP4 levels were lower in A549/CKAP4 KO/CKAP4C100S-HA cells when compared with A549/CKAP4 KO/CKAP4WT-HA cells (Figure 3A, Exosome). Exosomal CKAP4 had accumulated in the CM in a time-dependent manner until 48 h. The delayed accumulation was observed in A549/CKAP4 KO/CKAP4C100S-HA cells when compared with A549/CKAP4 KO/CKAP4WT-HA cells, although the exosomal marker clathrin was detected at the same levels in both groups (Figure 3B). Degradation velocities were also similar between the groups (Figure S3B). Typically, internalized proteins are trafficked to early endosomes and sorted to different pathways, e.g., delivered to recycling endosomes, lysosomes, and MVBs (31). Palmitoylation was non-essential for CKAP4 trafficking from the Golgi to the PM, and also DKK1-induced internalization in pancreatic S2-CP8 cells (25). WT-CKAP4 was internalized after DKK1 stimulation and reappeared in the PM approximately 3–4 h later; CKAP4C100S was also recycled but its recycling velocity was slower than WT-CKAP4 (Figure 3C), suggesting that palmitoylation promoted CKAP4 recycling. To isolate lysosomal proteins, A549/CKAP4 KO/CKAP4WT-HA and A549/CKAP4 KO/CKAP4C100S-HA cells were biotinylated and subjected to MACS (Figure 3D,3E). The collection of lysosomes and PM fractions was confirmed by the presence of lysosomal marker protein LAMP1 and low density lipoprotein receptor (LDLR), a cell surface protein which is transfer to the lysosome after endocytosis for degradation (33) (Figure 3F). Biotinylated lysosomal proteins, which represented cell surface proteins internalized from the PM, were recovered by streptoavidin beads (AP) and were significantly abundant in A549/CKAP4 KO/CKAP4C100S-HA cells when compared with A549/CKAP4 KO/CKAP4WT-HA cells (Figure 3F). Therefore, non-palmitoylated CKAP4 tended to be trafficked to the lysosome, thereby decreasing CKAP4 quantities secreted with exosomes.
An anti-CKAP4 antibody inhibits lung cancer cell proliferation
In our previous study (11), when DKK1 or CKAP4 were depleted in A549 cells, AKT activity and cell proliferation were suppressed. Since these were knockdown studies, the biological significance of CKAP4 expression in lung cancer was examined in a gain-of-function manner in NCI-H292 cells, where DKK1 was highly expressed but CKAP4 poorly expressed (Figure 1A). CKAP4 was present in the PM and AKT was activated in H292 cells ectopically expressing CKAP4-HA (H292/CKAP4-HA cells) (Figure 4A). Consistently, CKAP4-HA expression promoted 2-dimension (2D) cell growth and sphere formation in NCI-H292 cells (Figure 4B,4C). The anti-CKAP4 antibody inhibited AKT activity in A549 but not in NCI-H292 cells. However, the anti-CKAP4 antibody clearly suppressed AKT activity and sphere formation in NCI-H292/CKAP4-HA cells (Figure 4C,4D).
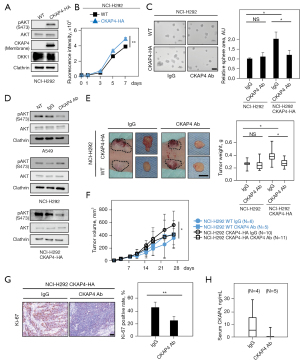
The role of CKAP4 in in vivo tumorigenesis of NCI-H292 cell was investigated by subcutaneously implanting cancer cells into immunodeficient mice. The volume and weight of xenograft tumors derived from NCI-H292/CKAP4-HA cells were significantly higher than tumors from WT cells (Figure 4E,4F). Moreover, xenograft tumors from NCI-H292/CKAP4-HA cells were decreased by the intraperitoneal injection of the anti-CKAP4 antibody, although the antibody did not affect WT-NCI-H292 cell tumor growth (Figure 4E,4F). Ki-67 positivity rates of the xenograft tumors of NCI-H292/CKAP4-HA cells were decreased by the anti-CKAP4 antibody (Figure 4G). Furthermore, the anti-CKAP4 antibody decreased CKAP4 serum levels in immunodeficient mice implanted with NCI-H292/CKAP4-HA cells (Figure 4H). These results indicated that CKAP4 localization in the PM promotes tumorigenesis in lung cancer cells and anti-CKAP4 antibody has therapeutic value on lung cancer cells activating DKK1-CKAP4 axis.
Anti-CKAP4 antibody and osimertinib combinations strengthen anti-tumor effects
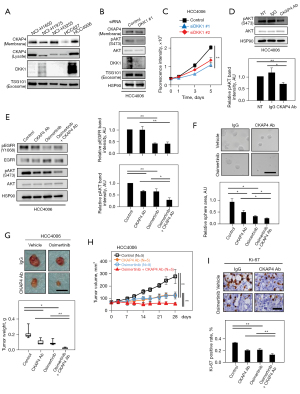
Osimertinib is a third-generation TKI and overcomes resistance mediated by EGFRT790M (34,35). To test the combination effects of the anti-CKAP4 antibody and osimertinib, DKK1 and CKAP4 expression levels were examined in NSCLC cell lines expressing EGFR mutations (Table S6). DKK1 was highly expressed and CKAP4 was present in the PM of HCC4006 cells which harbored the L747-E749del mutation (Figure 5A). DKK1 knockdown reduced AKT activity and cell proliferation in 2D cultures (Figure 5B,5C), and the anti-CKAP4 antibody decreased AKT activity in HCC4006 cells (Figure 5D). Osimertinib, but not the anti-CKAP4 antibody, inhibited EGFR activity, while both therapies suppressed AKT activity (Figure 5E). However, the combined anti-CKAP4 antibody and osimertinib strongly inhibited AKT activity, sphere formation, and xenograft tumor formation in HCC4006 cells when compared with monotherapy (Figure 5E-5H). Ki-67 positivity rates were more decreased in xenograft tumors treated with combined anti-CKAP4 antibody and osimertinib when compared with monotherapy (Figure 5I). Thus, the anti-CKAP4 antibody appears to exhibit an enhanced effect on NSCLC patients, activating both EGFR and DKK1-CKAP4 pathways, when combined with a TKI.
Discussion
The DKK1-CKAP4 axis is recognized as a cell proliferation signal in a subset of cancers (14,15). Our findings showed that: (I) CKAP4 was secreted with exosomes from NSCLC cells, (II) serum CKAP4 levels were higher in NSCLC patients than HCs, (III) CKAP4 palmitoylation controlled CKAP4 secretion with exosomes, and (IV) combination therapy with an anti-CKAP4 monoclonal antibody and osimertinib exhibited stronger anti-tumor effects in an in vivo mouse model.
Serum CKAP4 predicts CKAP4 expression in NSCLC
Exosomes are potential biomarkers for cancer diagnostics (36). CKAP4 serum levels were reportedly higher in lung cancer and esophageal squamous cell cancer patients than HCs (22,37). Consistent with these previous reports, our data also showed that higher CKAP4 values were identified more frequently in sera from NSCLC patients than HCs. Additionally, we observed that among 18 CKAP4 serum positive patients, 17 (94%) were immunohistochemically CKAP4 positive, suggesting that CKAP4 serum positivity reflected CKAP4 expression in tumors. We also observed that CKAP4 serum levels in all patients decreased after surgical resection of the primary lesion, indicating that serum CKAP4 could be a valuable follow-up marker in CKAP4 positive NSCLC cases.
It is important to identify eligible patients for molecular targeted drugs with respect to cost-effectiveness and loss of treatment opportunity. Our previous observations suggested that CKAP4 serum levels are higher in unresectable pancreatic cancer cases than resectable cases (19). Thus, CKAP4 expression in NSCLC tissue may be predicted from unresectable lung cancer cases without tumor biopsy that require invasive procedures; therefore, CKAP4 serum levels could be used to estimate CKAP4 expression in NSCLC and rapidly identify responders for anti-CKAP4 antibody therapy.
Palmitoylation enhances CKAP4 secretion with exosomes
Internalized CKAP4 is trafficked via two routes: (I) a recycling pathway to the PM via recycling endosomes for re-use as a receptor, and (II) an MVB pathway for release into the extracellular space with SEVs, including exosomes (19,25). In the present study, we established a new endocytotic pathway component fractionation method using MACS and dextran coated ferrofluid. By purifying lysosomes from cultured cells, we showed that CKAP4 is also trafficked to the lysosome and that palmitoylation controls CKAP4 levels trafficked to this organelle. Approximately 70% of endogenous CKAP4 in A549 cell PMs was palmitoylated. Since the CKAP4 extracellular region contributes to oligomer formation (6,38), palmitoylated and non-palmitoylated CKAP4 would form complexes in lipid rafts. Thus, a correct balance between palmitoylated and non-palmitoylated CKAP4 and their associated complexes may be important for regulating their microdomain localization. While it is unclear how palmitoylation affects CKAP4 secretion, one possibility is that CKAP4 interacts with specific molecular components responsible for cargo sorting in a lipid-dependent manner. Palmitoylated CKAP4 may prevent CKAP4-containing vesicles from moving to the lysosome or promote the vesicles to the MVB from early endosomes.
The anti-CKAP4 antibody is a promising therapy for lung cancer
Activating mutations in EGFR account for approximately 50% of all the mutations in lung adenocarcinoma in Asian population and several EGFR-TKI treatments have been developed for NSCLC patients harboring exon deletion, insertion or point mutations in EGFR (35). However, TKIs show approximate 70% response rates, some patients do not have specific molecular targets (35). Combination therapies are alternative treatment strategies that potentially address or delay resistance onset to EGFR-TKIs (39); however, they remain under investigation.
It was reported that hepatocellular carcinoma cells with high CKAP4 levels are featured by low proliferation capability and that CKAP4 associates with EGFR and suppresses the activation of EGFR signaling (40). However, the correlation between CKAP4 and EGFR in lung cancer has not been reported so far. Therefore, the correlation between CKAP4 and EGFR was analyzed based on TCGA dataset [Mixed Lung Adenocarcinoma (2022-v32)-tcga-589-tpm-gencode36] using the R2 Genomics Analysis and Visualization Platform. There was no statistically significant correlation between the transcription levels of CKAP4 and EGFR (R=0.043, P=0.297). Whether CKAP4 directly affects EGFR signaling in lung cancer is currently under way.
In principle, the concomitant use of osimertinib and other agents is currently not recommended (41). However, as resistance is an unsolved clinical challenge, new therapeutic strategies are urgently required. Previously, an anti-CKAP4 monoclonal antibody (3F11-2B10) exhibited tumor suppressive effects in pancreatic cancer S2-CP8 and HPAF-II cells, which express both DKK1 and CKAP4 (19). By overexpressing CKAP4 in NCI-H292 cells, both AKT activation and tumor growth in vitro and in vivo were induced, and these phenotypes were suppressed by anti-CKAP4 antibody to the levels of WT NCI-H292 cells. In addition, CKAP4 serum levels in mice transplanted with NCI-H292/CKAP-HA cells were decreased upon anti-CKAP4 antibody administration. Taken together with A549 cell data (19), these results support that CKAP4 is an appropriate molecular target of lung cancer cells expressing DKK1 and CKAP4. It was suggested that the PI3K-AKT pathway activation is a convergent feature of acquired EGFR-TKI resistance (42), and the inhibition of the PI3K-AKT pathway may prevent the development of secondary resistance to osimertinib (43). The anti-CKAP4 antibody exhibited additive tumor growth inhibitory effects by suppressing AKT, which was independent of osimertinib. Thus, a combined EGFR-TKI and anti-CKAP4 antibody strategy could underpin the development of novel therapies for lung cancer.
Conclusions
In this study, comprehensive analysis using clinical data, pathologic specimen, and serum samples from 92 lung cancer cases revealed that CKAP4 represents a novel biomarker and molecular target for lung cancer, and also plays a role as a companion diagnostic to predict the activation of DKK1-CKAP4 signaling. In vivo model demonstrated that the combination therapy with an anti-CKAP4 antibody and osimertinib provides a new lung cancer therapeutic strategy.
Acknowledgments
This study was supported by Dr. Yuri Terao (CentMeRE, Graduate School of Medicine, Osaka University) who provided technical assistance in preparing MISSION TRC shRNAs.
Funding: The work was supported by Grants-in-Aid for Scientific Research (2016–2021) (No. 16H06374) to A.K., for Early-Career Scientists (2018–2019) (No. 18K15064) to H.K., for Early-Career Scientists (2020–2021) (No. 20K16330) and (2022–2023) (No. 22K15511) to R.S., for Scientific Research (C) (2020–2022) (No. 20K07311) to H.Y., for Early-Career Scientists (2020–2021) (No. 20K17746) to A.N. from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and also supported by the Project for Cancer Research And Therapeutic Evolution (P-CREATE) [2018] (18cm0106132h0001) and (20cm0106152h0002) (2019–2021) and Translational Research Program (22ym0126039h0002) (2021–2023) to A.K. from the Japan Agency for Medical Research and development, AMED, and by Integrated Frontier Research for Medical Science Division, Institute for Open and Transdisciplinary Research Initiatives, Osaka University to A.K.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-571/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-571/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-571/coif). AK serves as the President of Federation of Asia and Oceanian Biochemist and Molecular Biologist (FAOBMB) without pay. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The human specimen protocol was approved by the ethics review board of the Graduate School of Medicine, Osaka University, Japan (Nos. 10026, 13455, 13563-8, and 18528-8). All studies were performed in accordance with committee guidelines and regulations. Written informed consent was provided by all patients. All animal experiments in this study protocols were approved by the Animal Research Committee of Osaka University, Japan (No. 21-048-1), in compliance with the Guidelines for Animal Experiments of Osaka University for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Fuchs HE, et al. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33. [Crossref] [PubMed]
- Zheng M. Classification and Pathology of Lung Cancer. Surg Oncol Clin N Am 2016;25:447-68. [Crossref] [PubMed]
- Arbour KC, Riely GJ. Systemic Therapy for Locally Advanced and Metastatic Non-Small Cell Lung Cancer: A Review. JAMA 2019;322:764-74. [Crossref] [PubMed]
- Schweizer A, Ericsson M, Bächi T, et al. Characterization of a novel 63 kDa membrane protein. Implications for the organization of the ER-to-Golgi pathway. J Cell Sci 1993;104:671-83. [Crossref] [PubMed]
- Schweizer A, Rohrer J, Slot JW, et al. Reassessment of the subcellular localization of p63. J Cell Sci 1995;108:2477-85. [Crossref] [PubMed]
- Klopfenstein DR, Klumperman J, Lustig A, et al. Subdomain-specific localization of CLIMP-63 (p63) in the endoplasmic reticulum is mediated by its luminal alpha-helical segment. J Cell Biol 2001;153:1287-300. [Crossref] [PubMed]
- Shibata Y, Shemesh T, Prinz WA, et al. Mechanisms determining the morphology of the peripheral ER. Cell 2010;143:774-88. [Crossref] [PubMed]
- Klopfenstein DR, Kappeler F, Hauri HP. A novel direct interaction of endoplasmic reticulum with microtubules. EMBO J 1998;17:6168-77. [Crossref] [PubMed]
- Vedrenne C, Hauri HP. Morphogenesis of the endoplasmic reticulum: beyond active membrane expansion. Traffic 2006;7:639-46. [Crossref] [PubMed]
- Harada T, Sada R, Osugi Y, et al. Palmitoylated CKAP4 regulates mitochondrial functions through an interaction with VDAC2 at ER-mitochondria contact sites. J Cell Sci 2020;133:jcs249045. [Crossref] [PubMed]
- Kimura H, Fumoto K, Shojima K, et al. CKAP4 is a Dickkopf1 receptor and is involved in tumor progression. J Clin Invest 2016;126:2689-705. [Crossref] [PubMed]
- Kikuchi A, Matsumoto S, Sada R. Dickkopf signaling, beyond Wnt-mediated biology. Semin Cell Dev Biol 2022;125:55-65. [Crossref] [PubMed]
- Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene 2006;25:7469-81. [Crossref] [PubMed]
- Kikuchi A, Fumoto K, Kimura H. The Dickkopf1-cytoskeleton-associated protein 4 axis creates a novel signalling pathway and may represent a molecular target for cancer therapy. Br J Pharmacol 2017;174:4651-65. [Crossref] [PubMed]
- Kagey MH, He X. Rationale for targeting the Wnt signalling modulator Dickkopf-1 for oncology. Br J Pharmacol 2017;174:4637-50. [Crossref] [PubMed]
- Kajiwara C, Fumoto K, Kimura H, et al. p63-Dependent Dickkopf3 Expression Promotes Esophageal Cancer Cell Proliferation via CKAP4. Cancer Res 2018;78:6107-20. [Crossref] [PubMed]
- Shinno N, Kimura H, Sada R, et al. Activation of the Dickkopf1-CKAP4 pathway is associated with poor prognosis of esophageal cancer and anti-CKAP4 antibody may be a new therapeutic drug. Oncogene 2018;37:3471-84. [Crossref] [PubMed]
- Chen ZY, Wang T, Gan X, et al. Cytoskeleton-associated membrane protein 4 is upregulated in tumor tissues and is associated with clinicopathological characteristics and prognosis in hepatocellular carcinoma. Oncol Lett 2020;19:3889-98. [Crossref] [PubMed]
- Kimura H, Yamamoto H, Harada T, et al. CKAP4, a DKK1 Receptor, Is a Biomarker in Exosomes Derived from Pancreatic Cancer and a Molecular Target for Therapy. Clin Cancer Res 2019;25:1936-47. [Crossref] [PubMed]
- Yamabuki T, Takano A, Hayama S, et al. Dikkopf-1 as a novel serologic and prognostic biomarker for lung and esophageal carcinomas. Cancer Res 2007;67:2517-25. [Crossref] [PubMed]
- Qiao R, Zhong R, Chang Q, et al. Serum dickkopf-1 as a clinical and prognostic factor in non-small cell lung cancer patients with bone metastases. Oncotarget 2017;8:79469-79. [Crossref] [PubMed]
- Yanagita K, Nagashio R, Jiang SX, et al. Cytoskeleton-Associated Protein 4 Is a Novel Serodiagnostic Marker for Lung Cancer. Am J Pathol 2018;188:1328-33. [Crossref] [PubMed]
- Zhang J, Planey SL, Ceballos C, et al. Identification of CKAP4/p63 as a major substrate of the palmitoyl acyltransferase DHHC2, a putative tumor suppressor, using a novel proteomics method. Mol Cell Proteomics 2008;7:1378-88. [Crossref] [PubMed]
- Schweizer A, Rohrer J, Kornfeld S. Determination of the structural requirements for palmitoylation of p63. J Biol Chem 1995;270:9638-44. [Crossref] [PubMed]
- Sada R, Kimura H, Fukata Y, et al. Dynamic palmitoylation controls the microdomain localization of the DKK1 receptors CKAP4 and LRP6. Sci Signal 2019;12:eaat9519. [Crossref] [PubMed]
- Harada T, Yamamoto H, Kishida S, et al. Wnt5b-associated exosomes promote cancer cell migration and proliferation. Cancer Sci 2017;108:42-52. [Crossref] [PubMed]
- Fujii S, Matsumoto S, Nojima S, et al. Arl4c expression in colorectal and lung cancers promotes tumorigenesis and may represent a novel therapeutic target. Oncogene 2015;34:4834-44. [Crossref] [PubMed]
- Matsumoto S, Fujii S, Sato A, et al. A combination of Wnt and growth factor signaling induces Arl4c expression to form epithelial tubular structures. EMBO J 2014;33:702-18. [Crossref] [PubMed]
- Nakai W, Yoshida T, Diez D, et al. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci Rep 2016;6:33935. [Crossref] [PubMed]
- Mato JM, Pencev D, Vasanthakumar G, et al. Inhibitors of endocytosis perturb phospholipid metabolism in rabbit neutrophils and other cells. Proc Natl Acad Sci U S A 1983;80:1929-32. [Crossref] [PubMed]
- Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 2014;30:255-89. [Crossref] [PubMed]
- Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol 2014;29:116-25. [Crossref] [PubMed]
- Zelcer N, Hong C, Boyadjian R, et al. LXR regulates cholesterol uptake through Idol-dependent ubiquitination of the LDL receptor. Science 2009;325:100-4. [Crossref] [PubMed]
- Jänne PA, Yang JC, Kim DW, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689-99. [Crossref] [PubMed]
- Remon J, Steuer CE, Ramalingam SS, et al. Osimertinib and other third-generation EGFR TKI in EGFR-mutant NSCLC patients. Ann Oncol 2018;29:i20-7. [Crossref] [PubMed]
- Kalluri R. The biology and function of exosomes in cancer. J Clin Invest 2016;126:1208-15. [Crossref] [PubMed]
- Chen L, You C, Jin X, et al. Cytoskeleton-associated protein 4 is a novel serodiagnostic marker for esophageal squamous-cell carcinoma. Onco Targets Ther 2018;11:8221-6. [Crossref] [PubMed]
- Chavda B, Ling J, Majernick T, et al. Antiproliferative factor (APF) binds specifically to sites within the cytoskeleton-associated protein 4 (CKAP4) extracellular domain. BMC Biochem 2017;18:13. [Crossref] [PubMed]
- Cooper AJ, Sequist LV, Lin JJ. Third-generation EGFR and ALK inhibitors: mechanisms of resistance and management. Nat Rev Clin Oncol 2022;19:499-514. [Crossref] [PubMed]
- Li SX, Liu LJ, Dong LW, et al. CKAP4 inhibited growth and metastasis of hepatocellular carcinoma through regulating EGFR signaling. Tumour Biol 2014;35:7999-8005. [Crossref] [PubMed]
- Di Noia V, D'Aveni A, D'Argento E, et al. Treating disease progression with osimertinib in EGFR-mutated non-small-cell lung cancer: novel targeted agents and combination strategies. ESMO Open 2021;6:100280. [Crossref] [PubMed]
- Jacobsen K, Bertran-Alamillo J, Molina MA, et al. Convergent Akt activation drives acquired EGFR inhibitor resistance in lung cancer. Nat Commun 2017;8:410. [Crossref] [PubMed]
- Leonetti A, Sharma S, Minari R, et al. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer 2019;121:725-37. [Crossref] [PubMed]


